Abstract
Background
Since the number of known lncRNA-disease associations verified by biological experiments is quite limited, it has been a challenging task to uncover human disease-related lncRNAs in recent years. Moreover, considering the fact that biological experiments are very expensive and time-consuming, it is important to develop efficient computational models to discover potential lncRNA-disease associations.
Results
In this manuscript, a novel Collaborative Filtering model called CFNBC for inferring potential lncRNA-disease associations is proposed based on Naïve Bayesian Classifier. In CFNBC, an original lncRNA-miRNA-disease tripartite network is constructed first by integrating known miRNA-lncRNA associations, miRNA-disease associations and lncRNA-disease associations, and then, an updated lncRNA-miRNA-disease tripartite network is further constructed through applying the item-based collaborative filtering algorithm on the original tripartite network. Finally, based on the updated tripartite network, a novel approach based on the Naïve Bayesian Classifier is proposed to predict potential associations between lncRNAs and diseases. The novelty of CFNBC lies in the construction of the updated lncRNA-miRNA-disease tripartite network and the introduction of the item-based collaborative filtering algorithm and Naïve Bayesian Classifier, which guarantee that CFNBC can be applied to predict potential lncRNA-disease associations efficiently without entirely relying on known miRNA-disease associations. Simulation results show that CFNBC can achieve a reliable AUC of 0.8576 in the Leave-One-Out Cross Validation (LOOCV), which is considerably better than previous state-of-the-art results. Moreover, case studies of glioma, colorectal cancer and gastric cancer demonstrate the excellent prediction performance of CFNBC as well.
Conclusions
According to simulation results, due to the satisfactory prediction performance, CFNBC may be an excellent addition to biomedical researches in the future.
Similar content being viewed by others
Background
Recently, accumulating evidences have indicated that lncRNAs (Long non-coding RNAs) are involved in almost the entire cell life cycle through various mechanisms [1, 2] and participate in close relationships in the development of some human complex diseases [3, 4] such as the Alzheimer’s disease [5] and many types of cancers [6]. Hence, identification of disease-related lncRNAs is critical to the understanding of the pathogenesis of complex diseases systematically and may further facilitate the discovery of potential drug targets. However, since biological experiments are very expensive and time-consuming, it has become a hot topic to develop effective computational models to uncover potential disease-related lncRNAs. Up to now, existing computational models for predicting potential associations between lncRNAs and diseases can be roughly classified into two major categories. Generally, in the first category of models, biological information of miRNAs, lncRNAs or diseases will be adopted to identify potential lncRNA-disease associations. For example, Chen et al. proposed a prediction model called HGLDA based on the information of miRNAs, in which, a hypergeometric distribution test was adopted to infer potential disease related lncRNAs [7]. Chen et al. proposed a KATZ measure to predict potential lncRNA-disease associations by utilizing the information of lncRNAs and diseases [8]. Ping and Wang et al. proposed a method for identifying potential disease-related lncRNAs based on the topological information of known lncRNA-disease association network [9]. In the second category of models, multiple data sources will be integrated to construct all kinds of heterogeneous networks to infer potential associations between diseases and lncRNAs. For example, Yu and Wang et al. proposed a naïve Bayesian Classifier based probability model to uncover potential disease-related lncRNAs by integrating known miRNA-disease associations, miRNA-lncRNA associations, lncRNA-disease associations, gene-lncRNA associations, gene-miRNA associations and gene-disease associations [10]. Zhang et al. developed a computational model to discover possible lncRNA-disease associations through combining lncRNAs similarity, protein-protein interactions and diseases similarity [11]. Fu et al. presented a prediction model by considering the quality and relevance of different heterogeneous data sources to identify potential lncRNA-disease associations [12]. Chen et al. proposed a novel prediction model called LRLSLDA by adopting Laplacian Regularized Least Squares to integrate known phenome-lncRNAome network, disease similarity network and lncRNA similarity network [13].
In recent years, in order to solve the problem of scarce known associations between different objects, an increasing number of recommender systems have been developed to increase the reliability of association prediction based on collaborative filtering methods [14], which depend on prior disposals to predict user-item relationships. Up to now, some novel prediction models have been proposed successively, in which, recommender algorithms have been appended to identify different potential disease-related objects. For example, Lu et.al proposed a model called SIMCLDA to predict potential lncRNA-disease associations based on inductive matrix completion by computing Gaussian interaction profile kernel of known lncRNA-disease associations, disease-gene and gene-gene onotology associations [15]. Luo et al. modeled drug repositioning problem into a recommendation system to predict novel drug indications based on known drug-disease associations through utilizing matrix completion [16]. Zeng et.al developed a novel prediction model called PCFM by adopting the probability-based collaborative filtering algorithm to infer gene-associated human diseases [17]. Luo et al. proposed a prediction model named CPTL to uncover potential disease-associated miRNAs via transduction learning by integrating disease similarity, miRNA similarity and known miRNA-disease associations [18].
In this study, a novel Collaborative Filtering model called CFNBC for predicting potential lncRNA-disease associations is proposed on the basis of Naïve Bayesian Classifier, in which, an original lncRNA-miRNA-disease tripartite network is constructed first by integrating miRNA-disease association network, miRNA-lncRNA association network and lncRNA-disease association network, and then, considering the fact that the number of known associations between the three objects such as lncRNAs, miRNAs and diseases is very limited, an updated tripartite network is further constructed by applying a collaborative filtering algorithm on the original tripartite network. Thereafter, based on the updated tripartite network, we can predict potential lncRNA-disease associations through adopting the Naïve Bayesian Classifier. Finally, in order to evaluate the prediction performance of our newly proposed model, LOOCV is implemented for CFNBC based on known experimentally verified lncRNA-disease associations. As a result, CFNBC can achieve a reliable AUC of 0.8576, which is much better than that of previous classical prediction models. Moreover, case studies of glioma, colorectal cancer and gastric cancer demonstrate the excellent prediction performance of CFNBC as well.
Results
Leave-one-out cross validation
In this section, in order to estimate the prediction performance of CFNBC, LOOCV will be implemented based on known experimentally verified lncRNA-disease associations. During simulation, for a given disease dj, each known lncRNA related to dj will be left out in turns as the test sample, whereas all the remaining associations between lncRNAs and dj are taken as training cases for model learning. Thus, the similarity scores between candidate lncRNAs and dj can be calculated and all candidate lncRNAs can be ranked by predicted results simultaneously. As a result, the higher the candidate lncRNA is ranked, the better the performance of our prediction model will be. Moreover, the value of area under the receive operating characteristic (ROC) curve (AUC) can be further used to measure the performance of CFNBC. Obviously, the closer the AUC value is to 1, the better the prediction performance of CFNBC will be. Hence, by setting different classification thresholds, we can calculate the true positive rate (TPR or sensitivity) and the false positive rate (FPR or 1-specificity) as follows:
Here, TP, FN, FP and TN denote the true positives, false negatives, false positives and true negatives respectively. Specifically, TPR indicates the percentage of candidate lncRNAs with ranks higher than a given rank cutoff, and FPR denotes the percentage of candidate lncRNAs with ranks below the given threshold.
The effects of α
Based on the assumption that original common neighboring miRNA nodes shall deserve more credibility than recommended common neighboring miRNA nodes, a decay factor α is used to make our prediction model CFNBC work more effectively. In this section, in order to evaluate the effects of α to the predcition performance of CFNBC, we will implement a series of experiments to estimate its actual effects while α is set to different values ranging from 0.05 to 0.8. As shown in Table 1, it is easy to see that CFNBC can achieve the best prediction performance while α is set to 0.05.
Comparison with other state-of-the-art methods
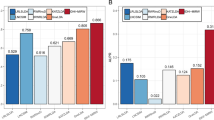
In order to further assess the performance of CFNBC, in this section, we will compare it with four kinds of state-of-the-art prediction models such as HGLDA [7], SIMLDA [15], NBCLDA [10] and the method proposed by Yang et al. [19] in the framework of LOOCV while α is set to 0.05. Among these four methods, since a hypergeometric distribution test was utilized to infer lncRNA-disease associations by integrating miRNA-disease associations with lncRNA-miRNA associations in HGLDA, then we will adopt a data set consisting of 183 experimentally validated lncRNA-disease associations as the hypergeometric distribution test to compare CFNBC with HGLDA. As illustrated in Table 2 and Fig. 1, the simulation results demonstrate that CFNBC outperforms HGLDA significantly. As for the model SIMLDA, since it applied inductive matrix completion to identify lncRNA-disease associations by integrating lncRNA-disease associations, gene-disease and gene-gene ontology associations, then we will collect a sub data set, which belongs to DSld in CFNBC and consists of 101 known associations between 30 different lncRNAs and 79 different diseases, from the data set adopted by SIMLDA to compare CFNBC with SIMLDA. As shown in Table 2 and Fig. 2, it is easy to see that CFNBC can achieve a reliable AUC of 0.8579, which is better than the AUC of 0.8526 achieved by SIMLDA. As for the model NBCLDA, since it fused multiple heterogeneous biological data sources and adopted the naïve Bayesian classifier to uncover potential lncRNA–disease associations, then we will compare CFNBC with it based on the data set DSld directly. As illustrated in Table 2 and Fig. 3, it is obvious that CFNBC can obtain a reliable AUC of 0.8576, which is higher than the AUC of 0.8519 achieved by NBCLDA as well. Finally, while comparing CFNBC with the method proposed by yang et al., in order to keep the fairness in comparison, we will collect a data set consisting of 319 lncRNA-disease associations between 37 lncRNAs and 52 diseases by deleting the nodes with degree equal to 1 on the data set DSld. As shown in Table 2 and Fig. 4, it is easy to see that CFNBC can achieve a reliable AUC of 0.8915, which considerably outperforms the AUC of 0.8568 achieved by the method proposed by yang et al. Hence, it is easy to draw a conclusion that our model CFNBC can achieve better performance than these classical prediction models.
Additionally, in order to further evaluate the prediction performance of CFNBC, we will compare it with above four models based on the predicted top-k associations by using F1-score measure. During simulation, we will randomly choose 80% of known lncRNA-disease associations as the training set, whereas all remaining known and unknown lncRNA-disease associations are taken as testing sets. Since the sets of known lncRNA-disease associations in these models are different, we will set different threshold k to compare them with CFNBC. As shown in Table 3, it is easy to see that CFNBC outperforms these four kinds of state-of-the-art models in terms of F1-score measure as well. Moreover, the paired t-test also demonstrates that the performance of CFNBC is significantly better than the prediction results of other methods in terms of the F1-scores (p-value < 0.05, as illustrated in Table 4).
Case studies
In order to further demonstrate the capability of CFNBC in inferring new lncRNAs related to a given disease, in this section, we will implement case studies of glioma, colorectal cancer and gastric cancer for CFNBC based on the data set DSld. As a result, the top 20 disease-related lncRNAs predicted by CFNBC have been confirmed by manually mining relevant literatures, and corresponding evidences are listed in the following Table 5. Additionally, among these three kinds of cancers chosen for case studies, the glioma is one of the most lethal primary brain tumors with a median survival of less than 12 months, and 6 out of 100000 people may have gliomas [20], hence it is important to find potential associations between glioma and dysregulations of some lncRNAs. As illustrated in Table 5, while applying CFNBC to predict candidate lncRNAs related to glioma, it is easy to see that there are six out of the top 20 predicted glioma-related lncRNAs having been validated by recent literatures on biological experiments. For instance, the lncRNA XIST has been demonstrated to be an important regulator in tumor progression and may be a potential therapeutic target in the treatment of glioma [21]. Ma et al. found that the lncRNA MALAT1 plays an important role in glioma progression and prognosis and may be considered as a convictive prognostic biomarker for glioma patients [22]. Xue et al. provided a comprehensive analysis of KCNQ1OT1-miR-370-CCNE2 axis in human glioma cells and a novel strategy for glioma treatment [23].
As for the colorectal cancer (CRC), it is the third most common cancer and the third leading cause of cancer death in men and women in the United States [24]. In recent years, accumulating evidences have shown that many CRC-related lncRNAs have been reported based on biological experiments. For example, Song et al. demonstrated that the higher expression of XIST was correlated with worse disease free survival of CRC patients [25]. Zheng et al. proved that the higher expression level of MALAT1 may serve as a negative prognostic marker in stage II/III CRC patients [26]. Nakano et al. found that the loss of imprinting of the lncRNA KCNQ1OT1 may play an important role in the occurrence of CRC [27]. As illustrated in Table 5, while applying CFNBC to uncover candidate lncRNAs related to CRC, it is obvious that there are 6 out of the top 20 predicted CRC-related lncRNAs having been verified in the Lnc2Cancer database.
Moreover, the gastric cancer is the second most frequent cause of cancer death [28]. Up to now, lots of lncRNAs have been reported to be associated with gastric cancer. For instance, XIST, MALAT1, SNHG16, NEAT1, H19 and TUG1 were reported to be upregulated in gastric cancer [29,30,31,32,33,34]. As illustrated in Table 5, while applying CFNBC to uncover candidate lncRNAs related to gastric cancer, it is obvious that there are 6 out of the top 20 newly identified lncRNAs related to gastric cancer having been validated by the lncRNADisease and Lnc2Cancer database respectively.
Discussion
Accumulating evidences have shown that prediction of potential lncRNA-disease associations is helpful in understanding crucial roles of lncRNAs in biological process, complex disease diagnoses, prognoses and treatments. In this manuscript, we constructed an original lncRNA-miRNA-disease tripartite network by combining miRNA-lncRNA, miRNA-disease and lncRNA-disease associations first. And then, we formulated the prediction of potential lncRNA-disease associations as a problem of recommender system and obtained an updated tripartite network through applying a novel item-based collaborative filtering algorithm to the original tripartite network. Finally, we proposed a prediction model called CFNBC to infer potential associations between lncRNAs and diseases by applying the naïve Bayesian Classifier on the updated tripartite network. Comparing with state-of-the-art prediction models, CFNBC can achieve better performs in terms of AUC values without entirely relying on known lncRNAs-disease associations, which means that CFNBC can predict potential associations between lncRNAs and diseases even as these lncRNAs and diseases are not in known data sets. Additionally, we implemented LOOCV to evaluate the prediction performance of CFNBC, and the simulation results showed that the problem of limited positive samples existed in state-of-the-art models has been significantly solved in CFNBC by the addition of collaborative filtering algorithm and the predictive accuracy has been improved by adopting the disease semantic similarity to infer potential associations between lncRNAs and diseases. Moreover, case studies of glioma, colorectal cancer and gastric cancer were implemented to further estimate the performance of CFNBC, and simulation results demonstrated that CFNBC could be a useful tool for predicting potential relationships between lncRNAs and diseases as well. Of course, despite the reliable experimental results achieved by CFNBC, there are still some biases in our model. For example, it is noteworthy that there are many other types of data that can be utilized to uncover potential lncRNA-disease associations, therefore, the prediction performance of CFNBC would be improved by the addition of more types of data. In addition, the results of CFNBC may be affected by the quality of datasets and the numbers of known lncRNA-disease relationships as well. Furthermore, successfully established models in the other computational fields would inspire the development of lncRNA-disease association prediction, such as microRNA-disease association prediction [35,36,37], drug-target interaction prediction [38] and synergistic drug combinations prediction [39].
Conclusion
Finding out lncRNA-disease relationships is essential for understanding human disease mechanisms. In this manuscript, our main contributions are as follows: (1) An original tripartite network is constructed by integrating a variety of biological information including miRNA-lncRNA, miRNA-disease and lncRNA-disease associations. (2) An updated tripartite network is constructed by applying a novel item-based collaborative filtering algorithm on the original tripartite network. (3) A novel prediction model called CFNBC is developed based on the naïve Bayesian Classifier and applied on the updated tripartite network to infer potential associations between lncRNAs and diseases. (4) CFNBC can be adopted to predict a potential disease-related lincRNA or an potential lncRNA-related disease without relying on any known lncRNA-disease associations. (5) A recommendation system is applied in CFNBC, which guarantees that CFNBC can achieve effective prediction results in condition of scarce known lncRNA-disease associations.
Data collection and preprocessing
In order to construct our novel prediction model CFNBC, we combined three kinds of heterogeneous data sets such as the miRNA-disease association set, the miRNA-lncRNA association set and the lncRNA-disease association set to infer potential associations between lncRNAs and diseases, which were collected from different public databases including the HMDD [40], the starBase v2.0 [41], and the MNDR v2.0 databases [42], etc.
Construction of the miRNA-disease and miRNA-lncRNA association sets
Firstly, we downloaded two datasets of known miRNA-disease associations and miRNA-lncRNA associations from the HMDD [40] in August 2018 and the starBase v2.0 [41] in January 2015 respectively. Then, we removed duplicated associations with conflicting evidences on these two data sets separately, manually picked out the common miRNAs existing in both the dataset of miRNA-disease associations and the dataset of miRNA-lncRNA associations, and retained only the associations related with these selected miRNAs in these two data sets. As a result, we finally obtained a data set DSmd including 4704 different miRNA-disease interactions between 246 different miRNAs and 373 different diseases, and a data set DSml including 9086 different miRNA-lncRNA interactions between 246 different miRNAs and 1089 different lncRNAs (see Supplementary Materials Table 1and Table 2).
Construction of the lncRNA-disease association set
Firstly, we downloaded a dataset of known lncRNA-disease associations from the MNDR v2.0 databases [42] in 2017. Then, once the dataset was collected, in order to keep the uniformity of disease names, we transformed some diseases names included in the set of lncRNA-disease associations into their aliases in the data set of miRNA-disease associations, and unified the names of lncRNAs in the datasets of miRNA-lncRNA associations and lncRNA-diseases associations. By this means, we selected out these lncRNA-disease interactions associated with both lncRNAs belonging to DSml and diseases belonging to DSmd. As a result, we finally obtained a data set DSld including 407 different lncRNA-disease interactions between 77 different lncRNAs and 95 different diseases (see Supplementary Materials Table 3).
Analysis of relational data sources
In CFNBC, the newly constructed lncRNA-miRNA-disease tripartite network (LMDN for abbreviation) consists of three kinds of objects such as lncRNAs, miRNAs and diseases. Therefore, we collected three kinds of relational data sources from different databases based on these three kinds of objects. As illustrated in Fig. 5, the numbers of diseases are 373 in the data set of miRNA-disease associations (m-d for abbreviation) and 95 in the data set of lncRNA-disease associations (l-d for abbreviation) respectively. The numbers of lncRNAs are 1089 in the data set of miRNA-lncRNA associations (m-l for abbreviation) and 77 in l-d respectively. The numbers of miRNAs are 246 in both m-l and m-d. Moreover, it is clear that the set of 95 diseases in l-d is a subset of the set of 373 diseases in m-d, and the set of 77 lncRNAs in l-d is a subset of the set of 1089 lncRNAs in m-l.
Method
As illustrated in Fig. 6, our newly proposed prediction model CFNBC consists of the following four main stages:
-
Step1: As illustrated in Fig. 6(a), we can construct a miRNA-disease association network MDN, a miRNA-lncRNA association network MLN, and an lncRNA-disease association network LDN based on the data sets DSmd, DSml and DSld respectively.
-
Step2: As illustrated in Fig. 6(b), through integrating these three newly constructed association networks MDN, MLN, and LDN, we can further construct an original lncRNA-miRNA-disease association tripartite network LMDN.
-
Step3: As illustrated in Fig. 6(c), after applying the collaborative filtering algorithm on LMDN, we can obtain an updated lncRNA-miRNA-disease association tripartite network LMDN′.
-
Step4: As illustrated in Fig. 6(d), after appending the naïve Bayesian classifier to LMDN′, we can obtain our final prediction model CFNBC.
Flowchart of CFNBC. In the diagram, the green circles, blue squares, and orange triangles represent lncRNAs, diseases and miRNAs respectively. a construction of MDN, MLN and LDN; (b) construction of the original tripartite network LMDN and its corresponding adjacency matrix; (c) construction of the updated tripartite network LMDN′ and its corresponding adjacency matrix; (d) prediction of potential lncRNA-disease associations through applying the naïve Bayesian classifier on LMDN′
In the original tripartite network LMDN, owing to the sparse known associations between lncRNAs and diseases, for any given lncRNA node a and disease node b, it is obvious that the number of miRNA nodes that associate with both a and b will be very limited. Hence, in CFNBC, we designed a collaborative filtering algorithm for recommending suitable miRNA nodes to corresponding lncRNA nodes and disease nodes respectively. And then, based on these known and recommended common neighboring nodes, we can finally apply the Naïve Bayesian Classifier on LMDN′ to uncover potential lncRNA-disease associations.
Construction of LMDN
Let matrix \( {R}_{MD}^0 \) be the original adjacency matrix of known miRNA-disease associations and the entity \( {R}_{MD}^0\left({m}_k,{d}_j\right) \) denote the element in the kth row and jth column of \( {R}_{MD}^0 \), then there is \( {R}_{MD}^0\left({m}_k,{d}_j\right) \) =1 if and only if the miRNA node mk is associated with the disease node dj, otherwise, there is \( {R}_{MD}^0\left({m}_k,{d}_j\right) \) =0. In the same way, we can obtain the original adjacency matrix \( {R}_{ML}^0 \) of known miRNA-lncRNA associations as well, and in \( {R}_{ML}^0 \), there is \( {R}_{ML}^0\left({m}_k,{l}_i\right) \) =1 if and only if the miRNA node mk is associated with the lncRNA node li, otherwise, there is \( {R}_{ML}^0\left({m}_k,{l}_i\right) \) =0. Additionally, considering that a recommender system may involve various input data including users and items, therefore, in CFNBC, we will take lncRNAs and diseases as users, while miRNAs as items. Thereafter, as for these two original adjacency matrices \( {R}_{MD}^0 \) and \( {R}_{ML}^0 \) obtained above, since their row vectors are the same, it is easy to see that we can construct another adjacency matrix \( {R}_{ML D}^0=\left[{R}_{ML}^0,{R}_{MD}^0\right] \) by splicing \( {R}_{MD}^0 \) and \( {R}_{ML}^0 \) together. Moreover, it is obvious that the row vector of \( {R}_{MLD}^0 \) is exactly the same as the row vector in \( {R}_{MD}^0 \) or \( {R}_{ML}^0 \), while the column vector of \( {R}_{MLD}^0 \) consists of the column vector of \( {R}_{MD}^0 \) and the column vector of \( {R}_{ML}^0 \).
Applying the item-based collaborative filtering algorithm on LMDN
Since CFNBC is based on the collaborative filtering algorithm, then the relevance scores between lncRNAs and diseases predicted by CFNBC will depend on the common neighbors between these lncRNAs and diseases. However, owing to the scarce known lncRNA-miRNA, lncRNA-disease and miRNA-disease associations, the number of common neighbors between these lncRNAs and diseases in LMDN will be very limited as well. Hence, in order to improve the number of common neighbors between lncRNAs and diseases in LMDN, we will apply the collaborative filtering algorithm on LMDN in this section.
First, on the basis of \( \kern0.50em {R}_{MLD}^0 \) and LMDN, we can obtain a co-occurrence matrix Rm × m, in which, let the entity R(mk, mr) denote the element in the kth row and rth column of Rm × m, then there is R(mk, mr) =1 if and only if the miRNA node mk and the miRNA node mr share at least one common neighboring node (a lncRNA node or a disease node) in LMDN, otherwise, there is R(mk, mr) =0. Hence, a similarity matrix R′ can be calculated after normalizing Rm × m as follows:
Where ∣N(mk)∣ represents the number of known lncRNAs and diseases associated to mk in LMDN, that is, the number of elements with value equaling to 1 in the kth row of \( {R}_{MLD}^0 \), |N(mr)| represents the number of elements with value equaling to 1 in the rth row of \( {R}_{MLD}^0 \), and ∣N(mk) ∩ N(mr)∣ denotes the number of known lncRNAs and diseases associated with both mk and mr simultaneously in LMDN.
Next, for any given lncRNA node li and miRNA node mh in LMDN, if the association between li and mh is known already, then, for a miRNA node mt other than mh in LMDN, it is obvious that the higher the relevance score between mt and mh, the bigger the possibility that there may exist potential association between li and mt. Hence, we can obtain the relevance score between li and mt based on the similarities between miRNAs as follows:
Here, N(li) represents the set of neighboring miRNA nodes that are directly connected to li in LMDN, and S(K, mt − top) denote the set of top-K miRNAs that are most similar to mt in LMDN. \( {R}_t^{\prime } \) is a vector consisting of the tth row of R′. In addition, there is uit = 1 if and only if li is interacted with mt in ML, otherwise, there is uit =0.
Similarly, for any given disese node dj and miRNA node mh in LMDN, if the association between dj and mh is known already, then, for a miRNA node mt other than mh in LMDN, we can obtain the relevance score between dj and mt based on the similarities between miRNAs as follows:
Where N(dj) denotes the set of neighboring miRNA nodes that are directly connected to dj in LMDN. In addition, there is ujt =1 if and only if dj is interacted with mt in MD, otherwise, there is ujt =0.
Obviously, based on the similarity matrix R′ and the adjacency matrix \( {R}_{MLD}^0 \), we can construct a new recommender matrix \( {R}_{MLD}^1 \) as follows:
In particular, for a certain lncRNA node li or a disease node dj in LMDN, if there is a miRNA mk satisfying \( {R}_{MLD}^0\left({m}_k,{l}_i\right)=1 \) or \( {R}_{MLD}^0\left({m}_k,{d}_j\right)=1 \) in \( {R}_{MLD}^0 \), then, we will first sum up the values of all elements in the ith or jth column of \( {R}_{MLD}^1 \) respectively. Thereafter, we will obtain its average value \( \overline{p} \). Finally, if there is a miRNA node mθ in the ith or jth column of \( {R}_{MLD}^1 \) satisfying \( {R}_{MLD}^1\left({m}_{\theta },{l}_i\right)>\overline{p} \) or \( {R}_{MLD}^1\left({m}_{\theta },{d}_j\right)>\overline{p} \), then we will recommend the miRNA mθ to li or dj respectively. And in the same time, we will as well add a new edge between mθ and li or mθ and dj in LMDN separately.
For instance, according to Fig. 6 and the given matrix \( {R}_{MLD}^0=\left[\begin{array}{cc}\begin{array}{cc}1& 1\\ {}1& 0\end{array}& \begin{array}{cc}1& 0\\ {}1& 0\end{array}\\ {}\begin{array}{cc}0& 1\\ {}\begin{array}{c}0\\ {}0\end{array}& \begin{array}{c}0\\ {}0\end{array}\end{array}& \begin{array}{cc}0& 1\\ {}\begin{array}{c}0\\ {}1\end{array}& \begin{array}{c}1\\ {}1\end{array}\end{array}\end{array}\right] \), we can obtain its corresponding matrices Rm × m, R′ and \( {R}_{MLD}^1 \) as follows:
To be specific, as illustrate in Figure 6, if taking the lncRNA node l1 as an example, then from the matrix \( {R}_{MLD}^0 \), it is easy to see that there are two miRNA nodes such as m1 and m2 associated with l1. In addition, according to formula (9), we can know as well that there is \( {R}_{MLD}^1\left({m}_5,{l}_1\right)=0.905>\overline{p}=\frac{R_{MLD}^1\left({m}_1,{l}_1\right)+{R}_{MLD}^1\left({m}_2,{l}_1\right)}{2}=\frac{0.81+0.81}{2}=0.81 \). Hence, we will recommend the miRNA node m5 to l1. In the same way, the miRNA nodes m2, m4 and m5 will be recommended to l2 as well. Moreover, according to previous description, it is obvious that these new edges between m5 and l1, m2 and l2, m4 and l2, and m5 and l2 will be added to the original tripartite network LMDN in the same time. Thereafter, we can obtain an updated lncRNA-miRNA-disease association tripartite network LMDN′ on the basis of the original tripartite network LMDN.
Construction of the prediction model CFNBC
The naïve Bayesian classifier is a kind of simple probabilistic classifier with a conditionally independent assumption. Based on this probability model, the posterior probability can be described as follows:
Where C is a dependent class variable and F1, F2, …, Fn are the feature variables of class C.
Moreover, since each feature Fi is conditionally independent to any other feature Fj (i ≠ j) in class C, then the above formula (10) can as well be expressed as follows:
In our previous work, we proposed a probability model called NBCLDA based on the Naïve Bayesian classifier to predict potential lncRNA-disease associations [10]. However, in NBCLDA, there exist some circumstances where it happens to be no relevance scores between a certain pair of lncRNA and disease nodes, and the reason is that there are no common neighbors between them owing to the scarce known associations between the pair of lncRNA and disease. Hence, in order to overcome this kind of drawback existing in our previous work, in this section, we will design a novel prediction model called CFNBC to infer potential associations between lncRNAs and diseases through adopting the item-based collaborative filtering algorithm on LMDN and applying the Naïve Bayesian classifier on LMDN′. In CFNBC, for a given pair of lncRNA and disease nodes, it is obvious that they will have two kinds of common neighboring miRNA nodes such as the original common miRNA nodes and the recommended common miRNA nodes. In order to illustrate this case more intuitively, an example is given in Figure 7, in which, the node m3 is an original common neighboring miRNA node since it has known associations with both l2 and d2, while the nodes m4 and m5 belong to recommended common neighboring miRNA nodes since they do not have known associations with both l2 and d2. And in particular, while applying the Naïve Bayesian classifier on LMDN′, for a given pair of lncRNA and disease nodes, we will consider that their common neighboring miRNA nodes, including both the original and recommended common neighboring miRNA nodes, are all conditionally independent of each other, since they are different nodes in LMDN′. That is, for a given pair of lncRNA and disease nodes, it is assumed that all their common neighboring nodes will not interfere with each other in CFNBC.
a subnetwork of Figure 6(d), in which, a solid line between a lcnRNA (or disease) node and a miRNA node means that there is a known association between these two nodes, while a dotted line between a lcnRNA (or disease) node and a miRNA node means that the association between these two nodes is obtained by our item-based collaborative filtering algorithm, then, it is easy to know that the common neighboring node m3 is an original common neighboring miRNA node of l2 and d2, while m4, m5 are recommended common neighboring miRNA nodes of l2 and d2
Method for applying the Naïve Bayesian theory on LMDN ′
For any given lncRNA node li and disease node dj in LMDN′, let CN1(li, dj) = {m1 − 1, m2 − 1, ⋯mh − 1} denote a set consisting of all original common neighboring nodes between them, and CN2(li, dj) = {m1 − 2, m2 − 2, ⋯mh − 2} denote a set consisting of all recommended common neighboring nodes between them in LMDN′, then, the prior probabilities \( p\left({e}_{l_i-{d}_j}=1\right) \) and \( p\left({e}_{l_i-{d}_j}=0\right) \) can be calculated as follows:
Where |Mc| denotes the number of known lncRNA-disease associations in LDN and |M| = nl × nd. Here, nl and nd represent the number of different lncRNAs and diseases in LDN respectively.
Furthermore, based on these two kinds of common neighboring nodes, the posterior probabilities between li and dj can be calculated as follows:
Obviously, comparing formula (14) with formula (15), it can be easily identified that whether an lncRNA node is related to a disease node or not in LMDN′. However, since it is too difficult to obtain the value of p(CN1(li, dj)) and p(CN2(li, dj)) directly, the probability of potential association existing between li and dj in LMDN′ can be defined as follows:
Here \( p\left({m}_{\updelta -1}|{e}_{l_i-{d}_j}=1\right) \) and \( p\left({m}_{\updelta -1}|{e}_{l_i-{d}_j}=0\right) \) denote the conditional possibilities that whether the node mδ − 1 is a common neighboring node between li and dj or not in LMDN′ separately, and \( p\left({m}_{\updelta -2}|{e}_{l_i-{d}_j}=1\right) \) and \( p\left({m}_{\updelta -2}|{e}_{l_i-{d}_j}=0\right) \) represent whether the node mδ − 2 is a common neighboring node between li and dj or not in LMDN′ respectively. Moreover, according to the Bayesian theory, these four kinds of conditional probabilities can be defined as follows:
Where \( p\left({e}_{l_i-{d}_j}=1|{m}_{\updelta -1}\right) \) and \( p\left({e}_{l_i-{d}_j}=0|{m}_{\updelta -1}\right) \) are the probability of whether the lncRNA node li is connected to the disease node dj or not respectively, while mδ − 1 is a common neighboring miRNA node between li and dj in LMDN′. And similarly, \( p\left({e}_{l_i-{d}_j}=1|{m}_{\updelta -2}\right) \) and \( p\left({e}_{l_i-{d}_j}=0|{m}_{\updelta -2}\right) \) represent the probability of whether the lncRNA node li is connected to the disease node dj or not respectively, while mδ − 2 is a common neighboring miRNA node between li and dj in LMDN′. Moreover, supposing that mδ − 1 and mδ − 2 are two common neighboring miRNA nodes between li and dj in LMDN′, let \( {N}_{m_{\updelta -1}}^{+} \) and \( {N}_{m_{\updelta -1}}^{-} \) represent the number of known associations and the number of unknown associations between disease nodes and lncRNA nodes in LMDN′ that have mδ − 1 as a common neighboring miRNA node between them, and \( {N}_{m_{\updelta -2}}^{+} \) and \( {N}_{m_{\updelta -2}}^{-} \) represent the number of known associations and the number of unknown associations between disease nodes and lncRNA nodes in LMDN′ that have mδ − 2 as a common neighboring miRNA node between them, then, it is obvious that \( p\left({e}_{l_i-{d}_j}=1|{m}_{\updelta -1}\right) \) and \( p\left({e}_{l_i-{d}_j}=1|{m}_{\updelta -2}\right) \) can be calculated as follows:
Obviously, according to above formula (17), formula (18), formula (19) and formula (20), the formula (16) can be modified as follows:
Furthermore, for any given lncRNA node li and disease node dj, since the value of \( \frac{p\left({e}_{l_i-{d}_j}=1\right)}{p\left({e}_{l_i-{d}_j}=0\right)} \) is a constant, then for convenience, we will denote the value of \( \frac{p\left({e}_{l_i-{d}_j}=1\right)}{p\left({e}_{l_i-{d}_j}=0\right)} \) as ϕm. In addition, for each common neighboring node mδ − 1 between li and dj, let Nl − 1 and Nd − 1 denote the numbers of lncRNAs and diseases associated to mδ − 1 in LMDN′ respectively, then it is obvious that there is \( {N}_{m_{\updelta -1}}^{+}+{N}_{m_{\updelta -1}}^{-}={N}_{l-1}\times {N}_{d-1} \). And similarly, for each common neighboring miRNA node mδ − 2 between li and dj, let Nl − 2 and Nd − 2 represent the numbers of lncRNAs and diseases associated to mδ − 2 in LMDN′ respectively, then it is obvious that there is \( {N}_{m_{\updelta -2}}^{+}+{N}_{m_{\updelta -2}}^{-}={N}_{l-2}\times {N}_{d-2} \). Thereafter, the above formula (16) can be further modified as follows:
Besides, since \( {N}_{m_{\updelta -1}}^{+} \) and \( {N}_{m_{\updelta -2}}^{+} \) may be zero, then we introduce the Laplace calibration to guarantee that the value of S(li, dj) will not be zero. Hence, the above formula (16) can once again be modified as follows:
Next, for any given lncRNA node and disease node, since the original common neighboring miRNA nodes between them are obtained from the known associations, while the recommended common neighboring miRNA nodes between them are obtained by our item-based collaborative filtering algorithm, then it is reasonable to consider that the original common neighboring miRNA nodes shall deserve more credibility than the recommended common neighboring miRNA nodes. Hence, in order to make our prediction model be able to work more effectively, we will add a decay factor α in the range of (0, 1) to the above formula (25). Thereafter, the formula (25) can be rewritten as follows:
Additionally, it has been reported that the degree of common neighboring nodes will play a significant role in the link prediction, and the common neighboring nodes with high degrees can improve the prediction accuracy [43]. Hence, we will further add an index Resource (RA) [44] and Logarithmic function for standardization to the above formula (26). Thereafter, for any given lncRNA node li and disease node dj in LMDN′, we can obtain the probability that there may exist a potential association between them as follows:
Here, \( {k}_{m_{\delta -1}} \) and \( {k}_{m_{\delta -2}} \) represent the degree of mδ − 1 and mδ − 2 in LMDN′ respectively.
Method for appending the disease semantic similarity into CFNBC
Each disease can be described as a Directed Acyclic Graph (DAG), in which, the nodes represent the disease MeSH descriptors and all MeSH descriptors in the DAG are linked from parent nodes to child nodes by a direct edge. By this way, a disease dj can be denoted as DAG(dj) = (dj, T(dj), E(dj)), where T(dj) is the set consisting of node dj and its ancestor nodes, E(dj) represents the set of edges between parent nodes and child nodes [45]. Thereafter, by adopting the scheme of DAG, we can define the semantic value of dj as follows:
Where,
Here, δ is the semantic contribution factor with the value between 0 and 1, and according to previous work, δ will be set to 0.5 in this paper. Thus, based on above formula (28) and formula (29), the semantic similarity between diseases dj and di can be calculated as follows:
Based on above formula (25) and formula (30), for any given lncRNA node li and disease node dj in LMDN′, we can finally obtain the probability that there may exist a potential association between them as follows:
Availability of data and materials
The Matlab code can be download at https://github.com/jingwenyu18/CFNBC;
The datasets generated and/or analysed during the current study are available in the HMDD repository, http://www.cuilab.cn/; MNDR repository, http://www.rna-society.org/mndr/; starBase repository, http://starbase.sysu.edu.cn/starbase2/index.php .
Abbreviations
- AUC:
-
areas under ROC curve
- CFNBC:
-
a novel Collaborative Filtering algorithm for sparse known lncRNA-disease associations will be proposed on the basis of Naïve Bayesian Classifier
- CRC:
-
the Colorectal cancer
- FPR:
-
false positive rates
- l-d :
-
the data set of lncRNA-disease associations
- LMDN:
-
the lncRNA-miRNA-disease tripartite network
- LMDN′:
-
an updated lncRNA-miRNA-disease association tripartite network
- lncRNA:
-
long non-coding RNAs lncRNA
- lncRNAs:
-
long non-coding RNAs lncRNAs
- LOOCV:
-
Leave-One Out Cross Validation
- m-d :
-
the data set of miRNA-disease associations
- m-l :
-
the data set of miRNA-lncRNA associations
- TPR:
-
true positive rates
References
Guttman MR, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154(1):240–51.
Guttman M, Rinn JL. Modular regulatory principles of large non–coding RNAs. Nature. 2012;482(7385):339–46.
Chen X, Yan CC, Zhang X, et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2016;18(4):558–76.
Chen X, Sun Y, Guan N, et al. Computational models for lncRNA function prediction and functional similarity calculation. Brief Funct Genomics. 2019;18(1):58–82.
Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat Med. 2008;14(7):723–30.
Li D, Liu X, Zhou J, et al. LncRNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of ERK and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2016;65(5):1612.
Chen X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep. 2015;5(1):13186.
Chen X. KATZLDA: KATZ measure for the lncRNA-disease association prediction. Sci Rep. 2015;5(1):16840.
Ping P, Wang L, Kuang L, et al. A novel method for LncRNA-disease association prediction based on an lncRNA-disease association network. IEEE/ACM Trans Comput Biol Bioinform. 2019;16(2):688–93.
Yu J, Ping P, Wang L, et al. A novel probability model for LncRNA-disease association prediction based on the Naïve Bayesian classifier. Genes. 2018;9(7):345.
Zhang J, Zhang Z, Chen Z, et al. Integrating multiple heterogeneous networks for novel LncRNA-disease association inference. IEEE/ACM Trans Comput Biol Bioinform. 2019;16(2):396–406.
Fu G, Wang J, Domeniconi C, et al. Matrix factorization-based data fusion for the prediction of lncRNA–disease associations. Bioinformatics. 2018;34(9):1529–37.
Chen X, Yan GY. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29(20):2617–24.
Liu NN, He L, Zhao M. Social temporal collaborative ranking for context aware movie recommendation. ACM Trans Intell Syst Technol. 2013;4(1):1–26.
Lu C, Yang M, Luo F, et al. Prediction of lncRNA-disease associations based on inductive matrix completion. Bioinformatics. 2018;34(19):3357–64.
Luo H, Li M, Wang S, et al. Computational drug repositioning using low-rank matrix approximation and randomized algorithms. Bioinformatics. 2018;34(11):1904–12.
Zeng X, Ding N, Rodríguez-Patón A, et al. Probability-based collaborative filtering model for predicting gene–disease associations. BMC Med Genet. 2017;10(Suppl 5):76.
Luo J, Ding P, Liang C, et al. Collective prediction of disease-associated miRNAs based on transduction learning. IEEE/ACM Trans Comput Biol Bioinform. 2017;14(6):1468–75.
Yang X, Gao L, Guo X, et al. A network based method for analysis of lncRNA-disease associations and prediction of lncRNAs implicated in diseases. PLoS One. 2014;9(1):e87797.
Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–710.
Wang Z, Yuan J, Li L, et al. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am J Transl Res. 2017;9(4):1845–55.
Ma KX, Wang HJ, Li XR, et al. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumor Biol. 2015;36(5):3355–9.
Gong W, Zheng J, Liu X, et al. Knockdown of long non-coding RNA KCNQ1OT1 restrained glioma cells’ malignancy by activating miR-370/CCNE2 axis. Front Cell Neurosci. 2017;11:84.
Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–17.
Song H, He P, Shao T, et al. Long non-coding RNA XIST functions as an oncogene in human colorectal cancer by targeting miR-132-3p. J buon. 2017;22(3):696–703.
Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–81.
Dong H, Xu G, Meng W, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7(16):22159–73.
Hartgrink HH, Jansen EP, Grieken NCV, et al. Gastric cancer. Lancet. 2009;374(9688):477–90.
Chen D, Ju H, Lu Y, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35(1):142.
Xia H, Chen Q, Chen Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7(35):56209–18.
Lian D, Amin B, Du D, et al. Enhanced expression of the long non-coding RNA SNHG16 contributes to gastric cancer progression and metastasis. Cancer Biomark. 2017;21(1):151–60.
Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142(7):1571–9.
Yang F, Bi J, Xue X, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279(17):3159–65.
Zhang E, He X, Yin D, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7(2):e2109.
Chen X, Xie D, Wang L, et al. BNPMDA: bipartite network projection for MiRNA-disease association prediction. Bioinformatics. 2018;34(18):3178–86.
Chen X, Huang L. LRSSLMDA:Laplacian regularized sparse subspace learning for MiRNA-disease association prediction. PLoS Comput Biol. 2017;13(12):e1005912.
Chen X, Huang L, Xie D, et al. EGBMMDA: extreme gradient boosting machine for MiRNA-disease association prediction. Cell Death Dis. 2018;9:3.
Chen X, Yan CC, Zhang X, et al. Drug-target interaction prediction: databases, web servers and computational models. Brief Bioinform. 2016;17(4):696–712.
Chen X, Ren B, Chen M, et al. NLLSS: predicting synergistic drug combinations based on semi-supervised learning. PLoS Comput Biol. 2016;12(7):e1004975.
Li Y, Qiu C, Tu J, et al. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42(D1):D1070–4.
Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–7.
Cui T, Zhang L, Huang Y, et al. MNDR v2. 0: an updated resource of ncRNA–disease associations in mammals. Nucleic Acids Res. 2017;46(D1):D371–4.
Zhou T, Lü L, Zhang Y, et al. Predicting missing links via local information. Eur Phys J B. 2009;71(4):623–30.
Liu W, Lü L. Link prediction based on local random walk. EPL (Europhysics Letters). 2010;89(5):58007.
Wang D, Wang J, Lu M, et al. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics. 2010;26(13):1644–50.
Acknowledgments
The authors thank the anonymous referees for suggestions that helped improve the paper substantially.
Funding
This research is partly sponsored by the National Natural Science Foundation of China (No.61873221, No.61672447) and the Natural Science Foundation of Hunan Province (No.2018JJ4058, No.2019JJ70010,No.2017JJ5036). Publication costs were funded by the National Natural Science Foundation of China (61873221, 61672447). The funder is Lei Wang (L.W.), whose contributions are stated in the section of Author’s Contributions.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.Y. and L.W.; Methodology, J.Y., Q.Z. and L.W.; Validation, Z.X., X.F. and Q.Z.; Formal Analysis, J.Y. and L.W.; Investigation, X.F. and Z.X.; Resources, Z.X. and Q.Z.; Data Curation, J.Y. and X.F.; Writing-Original Draft Preparation, J.Y. and Z.X; Writing-Review and Editing, L.W. and Q.Z.; Supervision, L.W.; Project Administration, L.W. and Q.Z.; Funding Acquisition, L.W. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests regarding the publication of this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yu, J., Xuan, Z., Feng, X. et al. A novel collaborative filtering model for LncRNA-disease association prediction based on the Naïve Bayesian classifier. BMC Bioinformatics 20, 396 (2019). https://doi.org/10.1186/s12859-019-2985-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-019-2985-0