Abstract
Background
The chloroplast signal recognition particle 54 (cpSRP54) is known for targeting the light-harvesting complex proteins to thylakoids and plays a critical role for chloroplast development in Arabidopsis, but little is known in rice. Here, we reported two homologous cpSRP54s that affect chloroplast development and plant survival in rice.
Results
Two rice cpSRP54 homologues, OscpSRP54a and OscpSRP54b, were identified in present study. The defective OscpSRP54a (LOC_Os11g05552) was responsible for the pale green leaf phenotype of the viable pale green leaf 14 (pgl14) mutant. A single nucleotide substitution from G to A at the position 278, the first intron splicing site, was detected in LOC_Os11g05552 in pgl14. The wild type allele could rescue the mutant phenotype. Knockout lines of OscpSRP54b (LOC_Os11g05556) exhibited similar pale green phenotype to pgl14 with reduced chlorophyll contents and impaired chloroplast development, but showed apparently arrested-growth and died within 3 weeks. Both OscpSRP54a and OscpSRP54b were constitutively expressed mainly in shoots and leaves at the vegetative growth stage. Subcellular location indicated that both OscpSRP54a and OscpSRP54b were chloroplast-localized. Both OscpSRP54a and OscpSRP54b were able to interact with OscpSRP43, respectively. The transcript level of OscpSRP43 was significantly reduced while the transcript level of OscpSRP54b was apparently increased in pgl14. In contrast, the transcript levels of OscpSRP54a, OscpSRP43 and OscpSRP54b were all significantly decreased in OscpSRP54b knockout lines.
Conclusion
Our study demonstrated that both OscpSRP54a and OscpSRP54b were essential for normal chloroplast development by interacting with OscpSRP43 in rice. OscpSRP54a and OscpSRP54b might play distinct roles in transporting different chloroplast proteins into thylakoids through cpSRP-mediated pathway.
Similar content being viewed by others
Background
Chloroplasts are the site for photosynthesis and other important metabolic processes such as fatty acid and amino acid biosynthesis (Nelson and Ben-Shem 2004; Lopez-Juez and Pyke 2005). The organelle contains up to several thousands of proteins, the majority of which are encoded in the nucleus with only a small fraction encoded in the plastid genome both in Arabidopsis and rice (Abdallah et al. 2000; Richly and Leister 2004). The nucleus-encoded chloroplast proteins are thus required to be transferred into the chloroplasts usually depending on the Toc/Tic complexes on the inner and outer chloroplast membranes (Jarvis and Robinson 2004; Schwenkert et al. 2011). Once these proteins enter into the chloroplast stroma, they may take on the final form, or further transport into the thylakoids through four distinct transport pathways namely, cpSec, ΔpH/Tat, cpSRP and spontaneous pathways (Schünemann 2007; Jarvis 2008).
The chloroplast signal recognition particle (cpSRP) and its receptor (cpFtsY) are involved in transporting of chloroplast proteins such as mature light-harvesting chlorophyll a/b binding proteins (LHCPs) to thylakoid membranes (Akopian et al. 2013). Defective or deficient in cpSRP and cpFtsY would impair the biogenesis of chloroplasts, leading to chlorophyll degeneration and chlorotic leaves in plants (Amin et al. 1999; Klimyuk et al. 1999; Asakura et al. 2004; Rutschow et al. 2008; Lv et al. 2015). In Arabidopsis, two null mutants of cpSRP54, ffc1–1 and ffc1–2, produce yellow first true leaves that become green subsequently. The levels of reaction center proteins are significantly lower in the young ffc1–2 leaves but recover to the normal protein level in adult plants grown on agar plates (Amin et al. 1999). However, the ffc1–2 plants are pale green with a fewer number of leaves and reduced rosette diameter grown under soil conditions (Rutschow et al. 2008). The null mutant of Arabidopsis cpSRP43, chaos, exhibits pale green leaves including the first true leaves in the whole lifecycle, with elevated chlorophyll a/b ratio, but a normal level of reaction center proteins (Amin et al. 1999; Klimyuk et al. 1999). The double mutant ffc/chaos exhibits pale yellow leaves at all growth stages with drastically reduced levels of LHCPs except Lhcb4 (Hutin et al. 2002). Similarly, the chlorophyll synthesis and chloroplast development are impaired in company with altered expression of chlorophyll synthesis-associated genes in rice OscpSRP43 mutants, w67 and pgl3, both of them show pale green leaves at all growth stages (Lv et al. 2015; Ye et al. 2018). Furthermore, high temperatures inhibit plant growth and facilitate the progression of leaf senescence in pgl3 (Ye et al. 2018). The maize cpFtsY mutant csr1–1 exhibits a pale yellow-green phenotype while csr1–3 shows a slight pale green phenotype (Asakura et al. 2004). Interestingly, both csr1–1 and csr1–2 are seedling lethal, similar to the Arabidopsis mutants cpFtsY-1 and cpFtsY-2, with completely arrested-growth at the cotyledon or the first true leaf stage under photoautotrophic conditions (Asakura et al. 2004; Asakura et al. 2008). It has been shown that the mutation lines of cpSRP pathway genes are able to accumulate truncated light-harvesting chlorophyll antenna (TLA) and enhance energy conversion efficiency in high-density cultures under bright sunlight conditions in Chlamydomonas reinhardtii (Kirst and Melis 2014; Jeong et al. 2017). Similar phenomena have been observed in tobacco, the cpSRP43 knockdown plants show lower chlorophyll contents in the RNAi canopy leaves with increased leaf-to-stem ratio, improved photosynthetic productivity and canopy biomass accumulation under high-density cultivation conditions (Kirst et al. 2018).
We previously identified a stable-inherited rice pale green leaf 14 (pgl14) mutant (originally termed HM14) at the vegetative stage (Shi et al. 2013). In this study, we isolated PGL14 that encoded for cpSRP54 (hereafter cpSRP54a). A single base substitution in the mutant allele resulting in an altered mRNA splicing is responsible for the pale green phenotype confirmed by genetic complementation. We also isolated a homologue of PGL14, LOC_Os11g05556 (hereafter cpSRP54b). Although the knockout lines of cpSRP54b displayed a similar pale green leaf phenotype to pgl14 with reduced chlorophyll levels, impaired chloroplast structures and down-regulated expression of chlorophyll synthesis/development related genes, they were seedling lethal. Both cpSRP54a and cpSRP54b were chloroplast-localized and could interact with OscpSRP43, respectively. Our results indicated that both of them are required for chloroplast development by interacting with cpSRP43 to potentially participate in protein transport into thylakoids in rice.
Results
Map-Based Isolation of PGL14
We previously identified a chlorophyll-deficient mutant pgl14 exhibiting pale green leaf phenotype from the first leaf to the flag leaf under natural conditions and mapped the recessive mutation to a 299 kb region in chromosome 11 (Shi et al. 2013). To fine map the mutation, a total of 1008 mutant-type F2 individuals derived from the cross pgl14/Moroberekan were used for genotyping. The PGL14 locus was further narrowed down to a 39.5 kb genomic region between RM26076 and RM26079, covering the BAC clones AC116949 and AC138169 (Fig. 1a). Seven open reading frames (ORFs) were annotated within this region in the database of Rice Genome annotation Project (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/), two of which (LOC_Os11g05552 and LOC_Os11g05556) were both annotated as hypothetical loci encoding for cpSRP54. Sequence analysis showed that a single nucleotide substitution from G to A at position 278 was detected in LOC_Os11g05552 in pgl14, and the mutation localized to the predicted splicing site on the last nucleotide of the first intron. RT-PCR analysis showed that the transcript in pgl14 was longer than that of WT, confirming the presence of the altered splicing transcript in the mutant (Fig. 1b). Sequence analysis also showed that the first intron of 119 bp was maintained in the mutant transcript which had a frame shift starting from valine at position 54 and terminating prematurely at position 131 (Fig. 2, Genbank accession MN105082). Therefore, LOC_Os11g05552 was most likely the candidate gene of PGL14.
Map-based cloning of PGL14.aPGL14 localizes to the short arm on chromosome 11 between RM26076 and RM26079, and is narrowed down to a 39.5 kb region covering the bacterial artificial chromosome clones AC116949 and AC138196. The 39.5 kb region contains 7 putative ORFs, the black box indicates LOC_Os11g05552, gray boxes indicate the other ORFs. LOC_Os11g05552 consists of 15 exons and 14 introns indicated as blank boxes and lines, respectively. The black arrow indicates the point mutation (G278A) at 1st intron splicing site. PTsF and PTsR are the forward and reverse primers for PCR analysis in (b). F and R are forward and reverse primers for qRT-PCR analysis; b RT-PCR shows different transcripts from WT and pgl14. The genomic DNA was used as a control
OscpSRP54a Rescues the Pale Green Phenotype
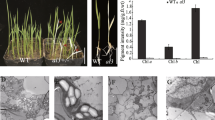
To verify the function of PGL14, the complementation vector cPGL carrying the entire coding region of PGL14, a 3.6 kb of upstream sequence and a 1.5 kb of downstream sequence was transformed into the rice calli induced from pgl14 mature embryo through Agrobacterium tumefaciens-mediated transformation. Five independent transgenic lines were obtained and showed the normal green phenotype similar to WT (Fig. 3a). Sequence characterization of these transgenic plants indicated that the complementary plants displayed a double peak (A and G) while pgl14 and WT presented a single peak, respectively (Fig. 3b), indicating that the wild-type allele has been incorporated into the mutant genome. Furthermore, RT-PCR analysis confirmed that both transcripts were presented in the transgenic plants (Fig. 3c). For the pigment levels, pgl14 showed apparently lowered Chl a, Chl b, carotenoid (Caro) and total chlorophyll (Chls) contents with significantly elevated Chl a/b ratio compared with WT, and all these parameters recovered to WT levels in complementary lines (Fig. 3d).. In addition, the chloroplast ultrastructure of complementary line C-pgl14 was similar to that of WT, displaying normal thylakoid membranes and stacked grana (Fig. 3e-g). Taken together, PGL14 was indeed the target gene responsible for the pale green leaf phenotype in pgl14, hereafter PGL14 is termed OscpSRP54a.
Functional complementation of OscpSRP54a. a Phenotype of WT, pgl14 and complementary line C-pgl14. Bar = 20 cm; b Sequence analysis of the mutation site (red arrow) in WT, pgl14 and C-pgl14; cOscpSRP54a transcripts in WT, pgl14 and C-pgl14; d Pigment contents in WT, pgl14 and C-pgl14 in 8-week-old leaves. Data are means ± SD (n = 3). Means with different letters indicate significant differences according to One-way ANOVA and Duncan’s test (p ≤ 0.01). Chloroplast ultrastructure of WT (e), pgl14 (f) and C-pgl14 (g) at the tillering stage. G, grana thylakoid; S, starch granule
cpSRP54s are Conserved in Plants
The cpSRP54 is widely present in photoautotrophic organisms. To simplify the phylogenetic analysis, we chose 11 OscpSRP54a homologues representing 10 species including the monocot, dicot and algae. Blastp analysis showed that OscpSRP54a shared 47–86% identity at the amino acid level with various cpSRP54 from Chlamydomonas reinhardtii (47%), Arabidopsis thaliana (70%), Glycine max (75%), Nicotiana tobacum (79%), Triticum urartu (81%), Zea mays (85%), Sorghum bicolor (86%), Chlorella sorokiniana (57%) and Spirulina subsalsa (55%) respectively (Fig. 4a). In addition, OscpSRP54a has 78% amino acid identity to OscpSRP54b which possess 47–78% amino acid identity with cpSRP54 from the other species. Apparently, the more closer relationship, the higher the amino acid identity shows in various cpSRP54s in which the function required for chloroplast development has been extensively elaborated in Arabidopsis and C. reinhardtii (Li et al. 1995; Yu et al. 2012; Jeong et al. 2017). The results indicated that cpSRPs were highly conserved in plant species.
Sequence comparison and phylogenetic tree of OscpSRP54a homologues. a Amino acid sequence comparison of OscpSRP54a homologues. Amino acid residues that are identical or similar are shaded in black and gray, respectively; b Phylogenetic tree of OscpSRP54a homologues including ZmcpSRP54 (Zea mays, XP_008679417), TucpSRP54 (Triticum urartu, EMS61888), GmcpSRP54 (Glycine max, XP_003521470), AtcpSRP54 (Arabidopsis thaliana, AAC64139), CrcpSRP54 (Chlamydomonas reinhardtii, AAK12834), NtcpSRP54 (Nicotiana tabacum, XP_016448785), SbcpSRP54 (Sorghum bicolor, KXG27759), OscpSRP54b (Oryza sativa L, ABG22368), CscpSRP54 (Chlorella sorokiniana, PRW50902), SscpSRP54 (Spirulina subsalsa, WP_017306040)
To reveal the evolutionary relationship of the cpSRP54 homologues, a phylogenetic tree was constructed. The results showed that cpSRP54 homologues in the higher plants could be classified into two groups as the monocot and dicot (Fig. 4b). Both OscpSRP54a and OscpSRP54b are clustered in the monocot group, however, OscpSRP54a is more closely related to TucpSRP54, ZmcpSRP54 and SbcpSRP54 than OscpSRP54b, probably indicating that OscpSRP54a and OscpSRP54b were likely originated differently.
OscpSRP54b is Essential for Chloroplast Development
To verify whether OscpSRP54b functions similar to OscpSRP54a, we introduced the CRISPR/Cas9 vectors cr1 and cr2 into Kitaake-derived embryogenic calli to edit the two specific target sites in exon 2 and exon 5, respectively (Fig. 5a). A total of 7 and 5 independent T0 transgenic lines were obtained from construct cr1 and cr2, respectively. Two (cr1–2 and cr1–5) out 7 T0 lines from construct cr1 were homozygous and exhibited pale green phenotype similar to pgl14 (Fig. 5b). Sequence analysis confirmed that there was one base deletion at the target site in cr1–2 and cr1–5, respectively (Fig. 5a). One (cr2–3) out of 5 T0 lines from construct cr2 was homozygous and showed pale green leaf phenotype similar to pgl14 (Fig. 5b), and sequence analysis showed that cr2–3 had a single nucleotide deletion at the target site (Fig. 5a). We then further characterized the performance of cr1–2 and cr2–3. Firstly, we determined the levels of photosynthetic pigments, and found that the contents of Chl a, Chl b, total Chls and carotenoid in cr1–2 and cr2–3 were significantly lower than those of Kitaake, and the Chl a/b ratio of both knockout mutants were much lower than in the WT (Fig. 5c). Then we observed the chloroplast ultrastructure by transmission electron microscopic analysis. The results showed that both cr1–2 and cr2–3 possessed a large number of hollow vesicles, reduced number of grana, irregular grana thylakoids, and destroyed stromal lamella compared with Kitaake (Fig. 5d-f). Moreover, cr1–2, cr2–3 as well as cr1–5 showed apparently arrested-growth and died within 3 weeks at the seedling stage. These results suggested that OscpSRP54b was essential for the chloroplast development and plant survival in rice.
Functional verification of OscpSRP54b for chloroplast development. a Deletion mutation at the target site in three representative knockout lines generated by the CRISPR/Cas9-mediated editing. cr1–2, cr1–5 and cr2–3 are homozygous mutants carrying 1-bp deletion on both homochromosomes. Black boxes indicate exons and lines indicate introns of OscpSRP54b. The sgRNA target sequence is underlined in blue and the PAM motif is highlighted in red letters, F and R are the forward and reverse primers for qRT-PCR analysis; b Phenotype of Kitaake and OscpSRP54b knockout mutants. Bar = 2 cm; c Pigment contents in 1 week-old leaves of Kitaake, cr1–2 and cr2–3. Different letters indicate significant differences according to One-way ANOVA and Duncan’s test (p ≤ 0.01); Chloroplast ultrastructure of Kitaake (d), cr1–2 (e) and cr2–3 (f). G, grana thylakoid; S, starch granule; OG, osmiophilic plastoglobuli; SL, stroma lamellae; HV, hollow vesicle
Rice cpSRPs are Mainly Expressed in Shoots and Leaves and cpSRPs Localize to Chloroplasts
To examine the expression pattern of OscpSRP54a and OscpSRP54b, qRT-PCR was carried out using samples from various tissues at the germination, tillering and heading stages. The results showed that the expressions of OscpSRP54a and OscpSRP54b were detected in all tissues tested with the highest expression levels of OscpSRP54a and OscpSRP54b in the leaves at the tillering stage, followed by the shoots at the germination stage (Fig. 6). Our results indicated that the expression patterns were similar between the two genes which were mainly expressed in above ground parts of the plants.
Spatial and temporal expression of OscpSRP54a and OscpSRP54b. a Relative expression levels of OscpSRP54a in various tissues of IR64 at different growth stages; b Relative expression levels of OscpSRP54b in various tissues of Kitaake at different growth stages. Different letters indicate significant differences according to One-way ANOVA and Duncan’s test (p ≤ 0.01)
To determine the subcellular location, we first predicted their physical locations using the ChloroP program (http://www.cbs.dtu.dk/services/ChloroP) and the results showed that both OscpSRP54a and OscpSRP54b were located in chloroplasts (Supplementary Fig. 1). The subcellular localization of OscpSRP54a and OscpSRP54b were then confirmed by expressing the constructs PAN580-OscpSRP54a and PAN580-OscpSRP54b in protoplasts. The green fluorescence signals of OscpSRP54a:GFP fusion protein and OscpSRP54b:GFP fusion protein overlapped with the chlorophyll autofluorescence signal whereas the free GFP signal was observed in the cytoplasm and nucleus (Fig. 7). These results demonstrated that both OscpSRP54a and OscpSRP54b were chloroplast-targeted proteins.
Down-Regulated Expression of OscpSRP43 in pgl14 and cr1–2
It has been shown that cpSRP43 interacts with cpSRP54 and is critical for chloroplast development by transporting proteins to thylakoids (Schünemann 2007; Akopian et al. 2013). To investigate whether the expression of cpSRP43 was affected in pgl14 and cr1–2, we measured the transcript levels of OscpSRP43, OscpSRP54a and OscpSRP54b by qRT-PCR. The results showed that the transcript level of OscpSRP43 was significantly decreased in pgl14 compared with WT, in contrast, the expression level of OscpSRP54b was significantly increased in pgl14 compared with WT, whereas the expression level of OscpSRP54a was similar between pgl14 and WT (Fig. 8a). In addition, the transcript levels of OscpSRP43, OscpSRP54a and OscpSRP54b were all notably reduced in cr1–2 compared with Kitaake (Fig. 8b). The results suggested that both mutations of cpSRP54a and cpSRP54b resulted in down-regulated expression of cpSRP43 in rice.
Differential expression of cpSRP genes. a Transcript levels of cpSRP genes in the wild type IR64 and pgl14 at 3 weeks after sowing. b Transcript levels of cpSRP genes in the wild type Kitaake and cr1–2 at 1 week after transplanting. The transcript levels of tested genes are normalized to the levels of Ubq (LOC_Os03g13170). The values are means ± SD (n = 3). The asterisk indicates the significant difference between the wild type and mutant according to Student’s t-test (* p ≤ 0.05; ** p ≤ 0.01)
The expression of genes associated with chlorophyll biosynthesis and chloroplast development was examined in pgl14 and cr1–2. The results showed that expression profile of most genes (such as PsbA, YGL1 and ChlD) was altered between the mutant and its wild type (Supplementary Fig. 2). These results further indicated that both OscpSRP54a and OscpSRP54b played an important role for normal development of chloroplasts in rice.
Both OscpSRP54a and OscpSRP54b Interact with cpSRP43 In Vivo
It has been shown that AtcpSRP54 interacts with AtcpSRP43 to form the heterodimer which binds to the L18 sequence of LHCPs to form the cpSRP-LHCPs complex for transporting LHCPs to thylakoids (Groves et al. 2001; Goforth et al. 2004). To verify whether OscpSRP54a and OscpSRP54b interact with OscpSRP43, the full length CDS of OscpSRP54a and OscpSRP54b were fused to C-terminal CFP respectively, and the full length CDS of OscpSRP43 was fused to N-terminal Venus. Co-expression of the OscpSRP54a-cCFP and OscpSRP43-nVenus fusion proteins in rice green tissue protoplasts produced obvious YFP signals overlapped with the auto fluorescence of chloroplasts (Fig. 9). The similar result was obtained by co-expression of OscpSRP54b-cCFP and OscpSRP43-nVenus fusion proteins. In contrast, co-expression of OscpSRP54a-cCFP and OsCSP41b-nVenus, or OscpSRP54b-cCFP and OsCSP41b-nVenus did not show the BiFC fluorescence (Fig. 9). Similarly, co-expression of OscpSRP54a-nVenus and OsCSP41b-cCFP, or OscpSRP54b-nVenus and OsCSP41b-cCFP did not show the BiFC fluorescence (Fig. 9). These results clearly demonstrated that both OscpSRP54a and OscpSRP54b interacted with OscpSRP43, respectively.
Discussion
Deficient chlorophyll contents shown by Arabidopsis cpSRP mutants indicate that cpSRP subunits play important roles for chloroplast development (Pilgrim et al. 1998; Amin et al. 1999; Hutin et al. 2002; Walter et al. 2015). Further studies on cpSRP43 and cpSRP54 defective mutants in Chlamydomonas and Arabidopsis suggest that the cpSRP43/SRP54 complex are able to recognize and bind to the hydrophobic LHCPs during passing through the stroma, thus, the dysfunctional cpSRP43 and/or cpSRP54 disrupt their assembly with LHCPs in thylakoids, leading to impaired chloroplast development (Amin et al. 1999; Hutin et al. 2002; Jeong et al. 2017). In the present study, we identified a chlorophyll-deficient mutant pgl14, which possessed a single nucleotide substitution at the splicing site of OscpSRP54a, leading to altered splicing transcripts and terminated prematurely. Complementation by the wild type allele could restore the pgl14 phenotype. OscpSRP54a is a homologue of Arabidopsis cpSRP54 with 70% identity at the amino acid level. The ffc mutants defective in AtcpSRP54 show severely yellow true leaves that subsequently become green (Amin et al. 1999). Unlike the ffc mutants, pgl14 exhibited pale green leaf phenotype in the whole life period. It is noticed that the rice mutant ygl138 has an 18 bp deletion in OscpSRP54 and shows a similar phenotype to pgl14, suggesting that ygl138(t) is allelic to PGL14 (Zhang et al. 2013). Unlike Arabidopsis which possesses only one copy of cpSRP54, the rice genome possesses two copies of cpSRP54s, OscpSRP54a and OscpSRP54b, which are physically adjacent and share 78% identity at the protein level. Independent knockout plants of OscpSRP54b were seedling lethal but showed pale green leaf phenotype and severely decreased chlorophyll content, resembling to pgl14. These results suggest that both OscpSRP54a and OscpSRP54b are indirectly associated with chlorophyll metabolism probably resulting from the impaired chloroplast development in the mutants.
It has been shown that AtcpSRP54 proteins are equally distributed between thylakoids and stroma by immunolocalization (Hutin et al. 2002). In the present study, we demonstrated that both OscpSRP54a and OscpSRP54b localized to chloroplasts, similar to OscpSRP43 (Lv et al. 2015). The thylakoid membranes in pgl14 seem to be disrupted, and the grana stacks were thinner than those of the wild-type at the tillering stage (Fig. 3e-g). Nevertheless, pgl14 is viable and capable of seeding at maturity (Shi et al. 2013). However, more severely destroyed stroma and a large number of hollow vesicles were found in the chloroplasts of the OscpSRP54b-knockout lines. The severe and irreversible chloroplast destruction could be the reason for the lethality of OscpSRP54b-knockout mutants. These results implicate that functional OscpSRP54b, but not OscpSRP54a, is necessary for rice survival.
Biochemical and genetic studies have indicated that cpSRP54 and cpSRP43 are able to form the heterodimeric complex to participate in localizing LHCPs to thylakoid membranes post-translationally (Hutin et al. 2002; Goforth et al. 2004; Dünschede et al. 2015). Our results suggested that both OscpSRP54a and OscpSRP54b could interact with OscpSRP43, respectively; indicating that OscpSRP54a and OscpSRP54b both functioned similarly as AtcpSRP54 for cpSRP-mediated protein transportation (Amin et al. 1999; Yu et al. 2012). Nevertheless, the existence of an alternative pathway has been confirmed in Arabidopsis that the targeting of LHCPs to thylakoids can be done when cpSRP54 is absent and unable to form the cpSRP-LHCP transit complex (Tzvetkova-Chevolleau et al. 2007). It has been shown that Glutamyl-tRNA reductase (GluTR) is the initial and rate-limiting enzyme for 5-aminolevolinic acid synthesis. Recent studies have demonstrated that cpSRP43 directly binds to GluTR and prevents aggregation of GluTR, thereby enhancing the stability of active GluTR (Wang et al. 2018). In pgl14 and cr1–2, the transcription levels of OscpSRP43 were significantly lower than those of the wild types, we speculated that downregulation of cpSRP43 in the mutants could lead to the decrease of stability and catalytic activity of GluTR, thus inhibiting the chlorophyll biosynthesis and ultimately leading to a decreased level of chlorophyll in the mutants. Interestingly, OscpSRP54b was significantly upregulated in pgl14, whereas OscpSRP54a was notably downregulated in cr1–2 mutant (Fig. 8). We speculate that OscpSRP54b might partially compensate for the defect of OscpSRP54a in pgl14, however, the severe destruction of chloroplasts in cr1–2 inhibited the transportation of LHCPs to thylakoids, resulting in the significantly down-regulation of both OscpSRP54a and OscpSRP54b.
It has been shown that the downregulation of genes involving in chlorophyll biosynthesis, photosynthesis and chloroplast development could be an indirect response to chlorophyll-deficient mutants (Yu et al. 2012; Lv et al. 2015; Qiu et al. 2018). For example, AtcpSRP54 mutations lead to decreased expression of AtGLK1, AtGLK2 and GUN4 which are related to plastid-to nucleus retrograde signaling (Lopez-Juez and Pyke 2005). In our study, the expression level of CHLH was significantly decreased in pgl14 and cr1–2. CHLH, a Mg-chelatase H subunit, is a multi-functional protein involved in plastid-to nucleus retrograde signaling and chlorophyll synthesis (Mochizuki et al. 2001; Jung et al. 2003; Wu et al. 2009; Tsuzuki et al. 2011). AtCpSRP54 is found to be associated with chloroplast ribosomes in the stroma, interacts with chloroplast synthesized thylakoid membrane proteins D1 and cytochrome b6 to perform its conserved role in co-translational targeting (Nilsson et al. 1999; Nilsson and vanWijk 2002; Piskozub et al. 2015). Similarly, PsbA encoding chloroplast D1 protein is significantly downregulated in pgl14 compared with the wild type. In contrast, the knockout mutant of OscpSRP54b induced a notable upregulation in PsbA transcription level. Studies in Synechocystis sp PCC 6803 have demonstrated that chlorophyll synthase/HliD complex binding with the Ycf39 protein and YidC/Alb3 insertase is involved in the photosystem II assembly, suggesting a link between chlorophyll biosynthesis and the Sec/YidC-dependent cotranslational insertion of nascent photosystem polypeptides into membranes (Chidgey et al. 2014; Knoppová et al. 2014). In our study, the expression level of YGL1, the homologue of Synechocystis sp PCC 6803 chlorophyll synthase in rice, was significantly downregulated in pgl14, but was significantly upregulated in cr1–2, these results implied that OscpSRP54b and OscpSRP54a might play distinct roles in transporting different chloroplast proteins into thylakoids through cpSRP-mediated pathway although the mechanism requires to be furthered studied.
Methods
Plant Materials
The pale green leaf 14 (pgl14) mutant was obtained from ethane methyl sulfonate (EMS) mutagenesis of the wild-type (WT) cultivar IR64 (Wu et al. 2005). The pale green phenotype is controlled by a single recessive nuclear gene (Shi et al. 2013). Normal green leaf cultivar Moroberekan was used as the male parent to cross with pgl14 for construction of an F2 fine mapping population. The parents and the population were grown under natural summer conditions in the paddy field at the China National Rice Research Institute (CNRRI), Hangzhou, China. The transgenic plants and WT were grown in the greenhouse at 30 °C and humidity of ca.70 with 16 h light/8 h dark cycle at CNRRI.
Measurement of Pigment Content
The total chlorophylls (Chl) were extracted from 10 mg fresh leaves with 95% alcohol in darkness for 48 h. The extracts were measured spectrophotometrically at 470 nm, 645 nm and 663 nm with a SpectraMax i3x Multi-Mode Microplate Reader (Molecular Devices, USA). Total Chl contents were determined according to the method of Arnon (Arnon 1949), and total carotenoid contents were determined as described by Wellburn (Wellburn 1994). All experiments were carried out with three biological replicates. Student’s t-test was conducted using EXCEL2013 and Duncan’s test was conducted by SAS 9.0. Means from three replicates were used for analysis.
Transmission Electron Microscopy Analysis
Full expanded leaves of pgl14, WT and C-pgl14 were collected at the seedling stage, while full expanded leaves of Kitaake, cpSRP54b knockout plants cr1–2 and cr2–3 were collected 1 week after transplanting. Leaf sections were fixed with 2.5% glutaraldehyde in phosphate buffer (pH 7.2) for 16 h at 4 °C, followed by rinsing, dewatering, embedding, and staining according to the method described by Lv et al. (2015). The chloroplast ultrastructure was observed by a Tecnai G2 F20 S-TWIN transmission electron microscope at Zhejiang University, Hangzhou, China.
Map-Based Cloning of PGL14 and Complementation Assay
The mutation was previously mapped to a 299 kb region in chromosome 11 (Shi et al. 2013). A total of 1008 mutant type F2 individuals derived from the cross pgl14/Moroberekan were used for fine mapping using simple sequence repeat (SSR) markers (Supplementary Table S1).
The genomic DNA was extracted following the minipreparation method (Lu and Zheng 1992). The genomic DNA fragments of the candidate gene were amplified from WT and pgl14, then sequenced and compared using DNASTAR software. RT-PCR analysis was used to confirm the splicing site in WT and pgl14 using the primers PTs (Supplementary Table S2). The sequences of the genomic DNA fragments and the transcripts were determined at Shanghai Invitrogen Inc. (Shanghai, China).
For functional complementation, a 9.6 kb WT genomic fragment containing a 4.5 kb entire open reading frame (ORF) of PGL14, a 3.6 kb upstream region, and a 1.5 kb downstream region was amplified using the specific primers PQf (Supplementary Table S2). The PCR products were double-digested with BamH I and Kpn I, and the fragments were recovered using the Axygen DNA gel extraction kit (Axygen scientific, USA). Then, the fragments were cloned into the binary vector pCAMBIA1300 to form a new transformation construct, cPGL. The new construct was introduced into the embryogenic calli generated from the mature seed embryos of pgl14 using Agrobacterium-mediated transformation method (Hiei and Komari 2008).
CRISPR/Cas9-Mediated Editing
To generate OscpSRP54b knockout mutants, two target sequences (TGGGCAGCTCACGACCGGGC, CCAACAGTTATCCTATTGG) were designed using CRISPR-P (Lei et al. 2014). The guide sequences were inserted into commercial gRNA expression vector VK005–01 respectively to create two new CRISPR/Cas9 vectors cr1 and cr2 (Supplementary Fig. 3) following the manufacture’s instruction (ViewSolid Biotech, Beijing). The cr1 and cr2 constructs were respectively introduced into Kitaake mature embryo-induced calli by Agrobacterium-mediated transformation (Hiei and Komari 2008).
Sequence Alignment and Phylogenetic Analysis
BlastP (https://www.ncbi.nlm.nih.gov/) was used to search homologous protein sequences of OscpSRP54a. The homologous sequences were aligned using the BioEdit software. The neighbor-joining phylogenetic tree was constructed using MEGA 5.1. 1000 bootstrap replicates were used for statistical support for the node values.
Quantitative Reverse Transcription PCR
Total RNA was extracted using the TRIzol method following the manufacture’s instruction (Invitrogen, USA). For RNA isolation, the roots and shoots of WT and Kitaake were collected from 10 day-old seedlings, the top full expanded leaves, leaf sheaths, roots, root stems, nodes and internodes of WT and Kitaake were collected from 10 week-old plants, roots, basals, nodes, internodes, flag leaves, flag leaf sheaths and panicles of WT and Kitaake were collected at the grain filling stage. For quantitative reverse transcription (qRT-PCR) analysis of genes associated with chlorophyll biosynthesis and chloroplast development, total RNA was extracted from the top full expanded leaves of pgl14 and WT at 3 weeks after sowing. Total RNA was extracted from 1 week-old leaves of cr1–2 and Kitaake. The first-strand cDNA was synthesized using the First Strand cDNA synthesis kit following the manufacturer’s protocol (TOYOBO Biotech, Japan). qRT-PCR was performed in a total volume of 20 μL qRT-PCR reaction buffer containing 2 μL reverse-transcribed product, 0.2 μM of each primer, and 2 × PowerUp SYBR Green PCR Master Mix (ThermoFisher Scientific, USA), on a Thermal Cycle Dice TM Real Time System II (Takara Biotech, Japan) with a cycling program of 2 m at 50 °C, 2 m at 95 °C, followed by 40 cycles of 15 s at 95 °C, 15 s at 55 °C, and 60 s at 72 °C. The ubiquitin gene (LOC_Os03g13170, Ubq) was used as an internal control. Primers used for qRT-PCR are listed in Supplementary Table S3. The means from three biological replicates were used for analysis by Student’s t-test and Duncan’s test by EXCEL2013 and SAS 9.0, respectively. The 2–ΔΔCT method was used to determine the relative transcript levels in gene expression.
Subcellular Localization and Bimolecular Fluorescence Complementation Assay
To determine the subcellular localization of OscpSRP54a and OscpSRP54b, their full length CDSs were amplified using the specific primers SLPGL14 and SLCr1, respectively (Supplement Table S2). The PCR products were double-digested with Xba I and BamH I, and the fragments were inserted into the 5′-terminal of GFP driven by the CaMV 35S promoter in the transient expression vector PAN580 to form the new constructs, PAN580-OscpSRP54a and PAN580-OscpSRP54b, respectively. For BiFC assay, the full length CDSs of OscpSRP54a and OscpSRP54b were amplified using the specific primers BiPGL14, and BiCr1. The full length CDS of OsCSP41b, which encodes for a chloroplast-localized protein (Mei et al. 2017), was amplified using the primer BiCSP41b and fused with cCFP and nVenus fragment as a control. The PCR products were double-digested with Kpn I and BamH I, and the fragments were inserted to the 5′-terminal of cCFP driven by the CaMV 35S promoter in the expression vector pE3449 to form three new constructs, OscpSRP54a-cCFP, OscpSRP54b-cCFP and OsCSP41b-cCFP, respectively. These fragments were inserted to the 5′-terminal of nVenus in pE3308 to form OscpSRP54a-nVenus, OscpSRP54b-nVenus and OsCSP41b-nVenus constructs. The full length CDS of OscpSRP43 was amplified using the primers BiW67 (Supplemental Table S2), and double-digested with Kpn I and Sma I, then the fragments were inserted to the 5′-terminal of nVenus driven by the 35S promoter in pE3308 to generate a new construct OscpSRP43-nVenus. The constructs were transformed into rice protoplasts according to the protocol described previously (Zhang et al. 2011).
Conclusions
OscpSRP54a and OscpSRP54b encode two homologous chloroplast signal recognition particles and their loss of function led to pale green leaves. Both OscpSRP54a and OscpSRP54b localize to the chloroplast and are able to interact with OscpSRP43, respectively. These results will facilitate efforts to further uncover the molecular mechanism of chloroplast protein transporting in monocots.
Availability of Data and Materials
Not applicable.
Abbreviations
- Chl:
-
Chlorophyll
- cpSRP:
-
Chloroplast signal recognition particle
- cpFtsY:
-
Chloroplast signal recognition particle receptor
- TLA:
-
Truncated light-harvesting chlorophyll antenna
- LHCPs:
-
Light harvesting chlorophyll a/b binding proteins
- CRISPR/Cas9:
-
Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9
References
Abdallah F, Salamini F, Leister D (2000) A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci 5:141–142
Akopian D, Shen K, Zhang X, Shan SO (2013) Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 82:693–721
Amin P, Sy DA, Pilgrim ML, Parry DH, Nussaume L, Hoffman NE (1999) Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol 121:61–70
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Asakura Y, Hirohashi T, Kikuchi S, Belcher S, Osborne E, Yano S, Terashima I, Barkan A, Nakai M (2004) Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16:201–214
Asakura Y, Kikuchi S, Nakai M (2008) Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J 56:1007–1017
Chidgey JW, Linhartová M, Komenda J, Jackson PJ, Dickman MJ, Canniffe DP, Koník P, Pilný J, Hunter CN, Sobotka R (2014) A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26:1267–1279
Dünschede B, Träger C, Schröder CV, Ziehe D, Walter B, Funke S, Hofmann E, Schünemann D (2015) Chloroplast SRP54 was recruited for posttranslational protein transport via complex formation with chloroplast SRP43 during land plant evolution. J Biol Chem 290:13104–13114
Goforth RL, Peterson EC, Yuan J, Moore MJ, Kight AD, Lohse MB, Sakon J, Henry RL (2004) Regulation of the GTPase cycle in post-translational signal recognition particle-based protein targeting involves cpSRP43. J Biol Chem 279:43077–43084
Groves MR, Mant A, Kuhn A, Koch J, Dübel S, Robinson C, Sinning I (2001) Functional characterization of recombinant chloroplast signal recognition particle. J Biol Chem 276:27778–27786
Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3:824–834
Hutin C, Havaux M, Carde JP, Kloppstech K, Meiherhoff K, Hoffman N, Nussaume L (2002) Double mutation cpSRP43−/cpSRP54- is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. Plant J 29:531–543
Jarvis P (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179:257–285
Jarvis P, Robinson C (2004) Mechanisms of protein import and routing in chloroplasts. Curr Biol 14:R1064–R1077
Jeong J, Baek K, Kirst H, Melis A, Jin E (2017) Loss of CpSRP54 function leads to a truncated light-harvesting antenna size in Chlamydomonas reinhardtii. Biochim Biophys Acta 1858:45–55
Jung KH, Hur J, Ryu CH, Choi Y, Chung YY, Miyao A, Hirochika H, An G (2003) Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol 44:463–472
Kirst H, Melis A (2014) The chloroplast signal recognition particle (CpSRP) pathway as a tool to minimize chlorophyll antenna size and maximize photosynthetic productivity. Biotechnol Adv 32:66–72
Kirst H, Shen Y, Vamvaka E, Betterle N, Xu D, Warek U, Strickland JA, Melis A (2018) Downregulation of the CpSRP43 gene expression confers a truncated light-harvesting antenna (TLA) and enhances biomass and leaf-to-stem ratio in Nicotiana tabacum canopies. Planta 248:139–154
Klimyuk VI, Persello-Cartieaux F, Havaux M, Contard-David P, Schuenemann D, Meiherhoff K, Gouet P, Jones JD, Hoffman NE, Nussaume L (1999) A chromodomain protein encoded by the Arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11:87–99
Knoppová J, Sobotka R, Tichy M, Yu J, Konik P, Halada P, Nixon PJ, Komenda J (2014) Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26:1200–1212
Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7:1494–1496
Li X, Henry R, Yuan J, Cline K, Hoffman NE (1995) A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc Natl Acad Sci U S A 92:3789–3793
Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49:557–577
Lu YJ, Zheng KL (1992) A simple method for DNA extraction in rice. Chin J Rice Sci 6:47–48 (in Chinese with English abstract)
Lv XG, Shi YF, Xu X, Wei YL, Wang HM, Zhang XB, Wu JL (2015) Oryza sativa chloroplast signal recognition particle 43 (OscpSRP43) is required for chloroplast development and photosynthesis. PLoS One 10:e0143249
Mei J, Li F, Liu X, Hu G, Fu Y, Liu W (2017) Newly identified CSP41b gene localized in chloroplasts affects leaf color in rice. Plant Sci 256:39–45
Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci U S A 98:2053–2058
Nelson N, Ben-Shem A (2004) The complex architecture of oxygenic photosynthesis. Nat Rev Mol Cell Biol 5:971–982
Nilsson R, Brunner J, Hoffman NE, vanWijk KJ (1999) Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J 18:733–742
Nilsson R, vanWijk KJ (2002) Transient interaction of cpSRP54 with elongating nascent chains of the chloroplast-encoded D1 protein; ‘cpSRP54 caught in the act’. FEBS Lett 524:127–133
Pilgrim ML, vanWijk KJ, Parry DH, Sy DA, Hoffman NE (1998) Expression of a dominant negative form of cpSRP54 inhibits chloroplast biogenesis in Arabidopsis. Plant J 13:177–186
Piskozub M, Króliczewska B, Króliczewski J (2015) Ribosome nascent chain complexes of the chloroplast-encoded cytochrome b6 thylakoid membrane protein interact with cpSRP54 but not with cpSecY. J Bioenerg Biomembr 47:265–278
Qiu ZN, Chen DD, He L, Zhang S, Yang ZN, Zhang Y, Wang ZW, Ren DY, Qian Q, Guo LB, Zhu L (2018) The rice white green leaf 2 gene causes defects in chloroplast development and affects the plastid ribosomal protein S9. Rice 11:39
Richly E, Leister D (2004) NUMTs in sequenced eukaryotic genomes. Mol Biol Evol 21:1081–1084
Rutschow H, Ytterberg AJ, Friso G, Nilsson R, vanWijk KJ (2008) Quantitative proteomics of a chloroplast SRP54 sorting mutant and its genetic interactions with CLPC1 in Arabidopsis. Plant Physiol 148:156–175
Schünemann D (2007) Mechanisms of protein import into thylakoids of chloroplasts. Biol Chem 388:907–915
Schwenkert S, Soll J, Bölter B (2011) Protein import into chloroplasts- how chaperones feature into the game. Biochim Biophys Acta 1808:901–911
Shi YF, Wei YL, Feng BH, Wang HM, Xu X, Huang QN, Lv XG, Zhang XB, Wu JL (2013) Genetic analysis and gene mapping of pale green leaf mutant HM14 in rice. Chin J Rice Sci 27:585–590 (in Chinese with English abstract)
Tsuzuki T, Takahashi K, Inoue S, Okigaki Y, Tomiyama M, Hossain MA, Shimazaki K, Murata Y, Kinoshita T (2011) Mg-chelatase H subunit affects ABA signaling in stomatal guard cells, but is not an ABA receptor in Arabidopsis thaliana. J Plant Res 124:527–538
Tzvetkova-Chevolleau T, Hutin C, Noël LD, Goforth R, Carde JP, Caffarri S, Sinning I, Groves M, Teulon JM, Hoffman NE, Henry R, Havaux M, Nussaume L (2007) Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19:1635–1648
Walter B, Pieta T, Schünemann D (2015) Arabidopsis thaliana mutants lacking cpFtsY or cpSRP54 exhibit different defects in photosystem II repair. Front Plant Sci 6:250
Wang P, Liang FC, Wittmann D, Siegel A, Shan SO, Grimm B (2018) Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis. Proc Natl Acad Sci U S A 115:E3588–E3596
Wellburn AR (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144:307–313
Wu FQ, Xin Q, Cao Z, Liu ZQ, Du SY, Mei C, Zhao CX, Wang XF, Shang Y, Jiang T, Zhang XF, Yan L, Zhao R, Cui ZN, Liu R, Sun HL, Yang XL, Su Z, Zhang DP (2009) The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol 150:1940–1954
Wu JL, Wu CJ, Lei C, Baraoidan M, Bordeos A, Madamba MR, Ramos-Pamplona M, Mauleon R, Potugal A, Ulat VJ, Bruskiewich R, Wang G, Leach J, Khush G, Leung H (2005) Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol Biol 59:85–97
Ye J, Yang YL, Wei XH, Niu XJ, Wang S, Xu Q, Yuan XP, Yu HY, Wang YP, Feng Y, Wang S (2018) PGL3 is required for chlorophyll synthesis and impacts leaf senescence in rice. J Zhejiang Univ Sci B 19:263–273
Yu B, Gruber MY, Khachatourians GG, Zhou R, Epp DJ, Hegedus DD, Parkin IA, Welsch R, Hannoufa A (2012) Arabidopsis cpSRP54 regulates carotenoid accumulation in Arabidopsis and Brassica napus. J Exp Bot 63:5189–5202
Zhang FT, Luo XD, Hu BL, Wan Y, Xie JK (2013) YGL138(t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 6:7
Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30
Acknowledgments
We thank Mr. Xiaotian Zhang from Jiangxi University of Finance and Economics for help in field sampling.
Funding
This study was financed by Technological Innovation Project of CAAS and the State Key Laboratory of Rice Biology (2017ZZKT10106).
Author information
Authors and Affiliations
Contributions
YS and YH performed most of the research and drafted the manuscript. XX and XZ carried out vector construction. XL carried out fine-mapping. YW and LL analyzed the data. JW supervised the study and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1 Supplementary Fig. 1
Subcellular localization prediction of OscpSRP54a and OscpSRP54b using ChloroP program. Supplementary Fig. 2 Expression of genes associated with chlorophyll biosynthesis and chloroplast development. Supplementary Fig. 3 Schematic structure of cr1 and cr2. Supplementary Table S1 Details of SSR markers for fine mapping of pgl14. Supplementary Table S2 Primer sequences for vector construction and reverse transcription-PCR. Supplementary Table S3 Primer sequences for RT-PCR.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, Y., He, Y., Lv, X. et al. Chloroplast SRP54s are Essential for Chloroplast Development in Rice. Rice 13, 54 (2020). https://doi.org/10.1186/s12284-020-00415-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-020-00415-2