Abstract
Background
Our previous studies indicated that miR-200b inhibits the growth of androgen-independent prostate cancer (AIPC) cells. In this study, we employed quantitative proteomics techniques to unravel the role of miR-200b in AIPC.
Methods
miR-200b was over-expressed or inhibited by transfection with miR-200b mimics or miR-200b inhibitor in PC3 cells. Total proteins were collected and the profiles of different groups were analyzed by label-free proteomics. PANTHER was applied to analyze biological processes, molecular functions and pathways of proteins regulated by miR-200b. miRBase was used to evaluate target genes of miR-200b in proteins regulated by miR-200b.
Results
Thirteen proteins were up-regulated in miR-200b mimics/mimics NC (negative miRNA control) and 14 proteins were down-regulated; 67 proteins were up-regulated in miR-200b inhibitor/inhibitor NC and 98 proteins were down-regulated. There were seven proteins which were both down-regulated by miR-200b mimics and up-regulated by miR-200b inhibitor, TM4SF1, YAP1, PPP1R2, MARCKS, RTN4, GLIPR2 and SUCLG1. Among these, TM4SF1, YAP1, PPP1R2, MARCKS, RTN4 were predicted as target genes of miR-200b by miRBase, while GLIPR2 and SUCLG1 were not.
Conclusion
This work identified several target genes of miR-200b by label free proteomics method, i.e., TM4SF1, YAP1, PPP1R2, MARCKS, RTN4, GLIPR2, and SUCLG1. The signaling pathways regulated by these proteins such as Hippo signaling may contribute to the phenotype resulting from miR-200b.
Similar content being viewed by others
Background
Prostate cancer (PCa) is one of the most common malignant tumors of the male genitourinary system and is the second most common cause of cancer-associated deaths in males [1]. Androgen deprivation therapy (ADT) is an effective therapeutic option for PCa. However, clinical outcomes of ADT in PCa patients have remained unsatisfactory because of castration resistance [2]. This type of prostate cancer is referred to as castration-resistant prostate cancer (CRPC) or androgen-independent prostate cancer (AIPC). Therefore, understanding the exact mechanisms that underlie androgen independence of PCa will help identify novel diagnostic biomarkers and facilitate the development of novel therapeutic strategies.
Small endogenous non-coding RNA (18–25 nt) are known as miRNAs. Accumulating evidence indicates that miRNAs play a critical role in AIPC. Previous studies have revealed that miR-200b are down-regulated in AIPC cells and that this phenomenon contributes to androgen-independent growth [3, 4]. Functional characterization of miRNAs depends strongly on identification of their specific mRNA binding partners because miRNAs can prevent protein expression by binding either the 3′UTR of the mRNA target through imperfect complementarity or via multiple sites by inhibition of the mRNA interaction with ribosomal complex and the translational machinery [5]. This kind of imperfect complementarity with target ensures that miRNAs have multiple intracellular targets that lead to amplification of the biological effects [6, 7]. Thus, the exact role of miR-200b in AIPC remains unclear although a subsequent study demonstrated that miR-200b mediates HIF1α-induced TXNDC5 expression under ADT conditions [8].
It is difficult to identify specific targets of miRNAs. Several tools for target prediction have been used to identify miRNA targets, such as TargetScan and microRNA.org (can be found in miRBase http://www.mirbase.org/), two of the most used tools. TargetScan and microRNA.org are user-friendly tools; however, there are too many potential targets in these databases. For example, there are 1193 predicted targets of miR-200b in TargetScan and 7176 in microRNA.org. In order to clarify the exact roles of miR-200b in AIPC, this study adopted a strategy to identify the target genes of miR-200b in AIPC using a proteomics method in combination with target prediction tools.
Methods
Cell culture
AIPC cell line PC3 and androgen-dependent prostate cancer (ADPC) cell line LnCap were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). A sub-line of LnCap named LnCap-AI, which was cultured in a phenol red-free RPMI 1640 with 10% FBS, depleted of steroids by charcoal/dextran-treatment (CDS medium) (Biological Industry, Israel), instead of regular FBS for more than nine passages. PC3 cells and LnCap-AI have been routinely used as in vitro models of androgen-independent stages of prostate cancers. In our previous study conducted on PC3 and LnCap-AI as a model system, down-regulation of miR-200b was found to contribute to the proliferation of AIPC [3]. Normally, these cells were cultured in 10% fetal bovine serum (FBS)-supplemented phenol red-containing RPMI 1640 (Gibco, Carlsbad, CA, USA), containing l-glutamine (2 mM) (Invitrogen, Carlsbad, CA, USA) and penicillin sodium (100 U/mL)/streptomycin sulphate (100 μg/mL) (Invitrogen, Carlsbad, CA, USA) in a humidified incubator at 37 °C with 5% CO2. For maintenance of androgen independence, the cell culture medium was changed to a phenol red-free RPMI 1640 with 10% FBS that was depleted of steroids by charcoal/dextran-treatment (CDS medium) (Biological Industry, Israel) instead of regular FBS.
RNA oligos and transfection
miR-200b mimics, negative miRNA control (mimics-NC), miR-200b inhibitor and negative inhibitor control (inhibitor-NC) were purchased from GenePharma (Shanghai, China). Cells in the exponential growth phase were plated at a density of 30–50% confluence, followed by incubation for 24 h. The cells were then transfected with small RNAs (0.05 μM) in reduced serum medium (OPTI-MEM-I), according to the manufacturer’s protocol.
Quantitative real-time PCR (Q-PCR) analysis for miRNA and mRNA expression
Forty-eight hours after RNA oligos transfection, miRNA and mRNA expressions were detected by Q-PCR. Total RNAs from cells were extracted using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instruction. Quantification of mature miRNAs and mRNA of target proteins was performed using the Quantitect SYBR Green PCR Kit (Stratagene, CA, USA) with a MX3005P multiplex quantitative PCR system (Stratagene, CA, USA), according to the manufacturer’s instructions. U6 small nuclear RNA was used as an internal control for determining the relative miRNA expression level; GAPDH was used as an internal control to determine the relative mRNA expression levels. The relative expression levels of miRNAs and mRNAs were calculated using the comparative ΔΔCT method [9, 10]. The fold-changes were calculated by the equation \( {2^{ - \Delta \Delta {C_{\rm{t}}}}} \). All primers used are listed in Additional file 1: Table S1.
Sample preparation for a label-free experiment
PC3 cells were divided into 4 groups and transfected with miR-200b mimics, miR-NC, miR-200b inhibitor and inhibitor-NC, respectively. Forty-eight hours after transfection, cell pellets were lysed, total proteins were collected and quantified with a BCA assay kit (Sigma-Aldrich, St. Louis, USA). Proteins (250 μg for each sample) were digested according to the FASP procedure described elsewhere [11]. The peptide content was estimated by UV light spectral density at 280 nm using an extinctions coefficient of 1.1 of 0.1% (g/L) solution, which was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins.
Liquid chromatography (LC)–electrospray ionization (ESI) tandem MS (MS/MS) analysis by Q exactive
The peptide of each sample was desalted on C18 Cartridges [Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 mL; Sigma-Aldrich, USA], then concentrated by vacuum centrifugation and reconstituted in 40 µL of 0.1% (v/v) trifluoroacetic acid. MS experiments were performed on a Q Exactive mass spectrometer that was coupled to Easy nLC (Thermo Fisher Scientific). 5 μg peptide was loaded onto a C18-reversed phase column (10 cm long, 75 μm inner diameter, 3 μm resin; Thermo Scientific, USA) in buffer A (2% acetonitrile and 0.1% formic acid) and separated with a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid). MS data were acquired using a data-dependent top 10 method, dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. Determination of the target value was based on predictive Automatic Gain Control (pAGC). MS experiments were performed in triplicate for each sample.
Sequence database searching and data analysis
The MS data were analyzed using MaxQuant software version 1.3.0.5. MS data were searched against the uniprot_human_152544_20160420.fasta (152544 total entries; downloaded April 20, 2016). An initial search was set at a precursor mass window of 6 ppm. The search employed an enzymatic cleavage rule of Trypsin/P and allowed maximal two missed cleavage sites and a mass tolerance of 20 ppm for fragment ions. Cysteine carbamidomethylation was defined as a fixed modification, while protein N-terminal acetylation and methionine oxidation were defined as variable modifications for database searching. Label-free quantification was carried out in MaxQuant, as described elsewhere [12]. The peptide spectrum match (PSM) was filtered by Posterior Error Probability (PEP). Max PEP is 0.1. The peptides were filtered based on andromeda score and false discovery rate (FDR). The cutoff level for andromeda score was set at 0 (andromeda score > 0). The cutoff level for FDR was set at 0.01 (peptide FDR < 0.01). Co-fragmentation generally reduces the number of peptides identified in database searches and poses special problems for reporter fragment based quantification methods because both peptides contribute to the measured ratios. Co-fragmentation was performed according to the algorithm of MaxQuant software [13]. Protein abundance was calculated on the basis of the normalized spectral protein intensity (LFQ intensity). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD006350.
Western blot
Forty-eight hours after RNA oligos transfection, target protein expressions were determined by Western blot. Cultured PC3 cells, treated under different conditions, were lysed by pre-chilled RIPA buffer. Total protein content was measured using a BCA (bicinchoninic acid assay) protein assay reagent kit. Lysates were mixed with Laemmli buffer and boiled for 5 min. 30 µg of protein were separated on a 12% SDS-PAGE gel followed by transfer to polyvinylidene difluoride (PVDF) membrane (Millipore), after which membranes were blocked with 5% non-fat dry milk in PBS/0.1% Tween-20 (PBST) solution. Membranes were immune-blotted overnight at 4 °C. Anti-PPP1R2 antibody (1:1000, ab58149; Abcam, USA), Anti-GLIPR2 antibody (1:250, ab122059; Abcam, USA), and Anti-MARCKS (D88D11) antibody (1:1000, #5607T; Cell Signaling Technology, USA) were used. An HRP-conjugated anti-rabbit antibody (Cell Signaling Technology, USA) was used as the secondary antibody. To confirm protein loading, membranes were probed with antibody recognizing GAPDH (1:1000). The Western blot data shown are representative for at least three independent experiments.
Gene enrichment analysis and functional annotation (PANTHER)
The PANTHER (protein analysis through evolutionary relationships) classification system was designed to classify proteins (and their genes) in order to facilitate high-throughput analysis. Proteins have been classified according to: family and subfamily, molecular function, biological process and pathway [14]. In order to maximize the coverage of important biological information, the proteomics identified proteins down regulated by miR-200b mimics and up-regulated by miR-200b inhibitor were subjected to enrichment analysis (beta) server (The PANTHER) for gene ontology analysis (http://geneontology.org/). The dataset was analyzed using the Core Analysis module to rank the proteins into top biological process (BP), molecular functions (MF) and pathway. Analysis type: PANTHER over-representation test (release 20160715); Annotation Version and Release Date: GO Ontology database Released 2016-06-22.
miR-200b targets prediction
The internet resource miRBase (http://www.miRBase.org/), the primary microRNA sequence repository [15], was used to predict potential miR-200b targets, using “hsa-miR-200b” as a search term.
KEGG pathway analysis
Hippo signaling pathway plays an important role in AIPC. In this study differentially expressed proteins from this pathway were analyzed with KEGG pathway database (http://www.kegg.jp/kegg/pathway.html).
Proteins interaction network analysis (STRING)
In this study, the intersection of proteins down-regulated by miR-200b mimics (fold ≥ 1.5, P < 0.05) and up-regulated by miR-200b inhibitor (fold ≥ 1.5, P < 0.05) were considered as candidates for the target protein list. Protein–protein interactions represent a crucial element of modern biological research fields such as proteomics study. The STRING knowledge database (http://string-db.org/) is a user-friendly and comprehensive database designed to assemble, evaluate and disseminate protein–protein association information [16, 17]. Subsequently, the candidate target gene list was imported into STRING for pathway analysis and network reconstruction.
Statistical analysis
All data are presented as mean ± SD. The differences between groups were analyzed by Significance B test of variance. P < 0.05 was considered statistically significant (two-tailed).
Results
Effect of miR-200b expression on the proliferation of prostate cells
The proliferation of PC3 and LnCap-AI cells with miR-200b over-expression or inhibition was detected by CCK-8. miR-200b over-expression decreased cell proliferation (P < 0.05), while miR-200b inhibition had no effect on proliferation (P > 0.05) of the PC3 and LnCap-AI cells cultured in phenol red-free RPMI 1640 with CDS (see our previous data [3]).
Identification of differentially-expressed proteins (DEPs) by label free proteomics
In this study, a total of 3792 proteins were identified in 4 samples by LC-ESI–MS/MS. To reduce false proteins detection, a global normalization method was applied for data quality control in order to minimize the variability due to sample preparation or equipment conditions.
Significance B test was applied to detect DEPs in miR-200b mimics/mimics NC and miR-200b inhibitor/inhibitor NC groups. Among these identified proteins, 13 proteins were up-regulated in miR-200b mimics/mimics NC and 14 proteins were down-regulated (fold ≥ 2, P < 0.05; Additional file 2: Table S2); 67 proteins were up-regulated in miR-200b inhibitor/inhibitor NC and 98 proteins were down-regulated (fold ≥ 2, P < 0.05; Additional file 3: Table S3).
Validation of the expression levels of DEPs by Q-PCR and Western blot
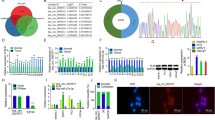
Only one protein, TM4SF1, was detected in both miR-200b mimics/mimics NC and miR-200b inhibitor/inhibitor NC groups using P value < 0.05 and a ratio of mean ≥ 2 or ≤ 0.5 as the criteria for significant up or down regulation. Thus, considering the efficiency of RNA oligos transfection, the intersection of proteins down-regulated by miR-200b mimics (fold ≥ 1.5, P < 0.05) and up-regulated by miR-200b inhibitor (fold ≥ 1.5, P < 0.05) were considered as the convincing target proteins. There were seven proteins in this list (Table 1, MS/MS spectra; see Additional file 4: Figs. S1–S7). The expressions of these seven proteins were determined by Q-PCR. The results showed that there were no significant changes in the expression of these proteins at the mRNA level following miR-200b mimics and miR-200b inhibitor transfection (Fig. 1b, Additional file 4: Fig. S8b). In order to verify the regulation of the proteins at the protein level, PPP1R2, MARCKS, and GLIPR2 were randomly chosen for validation by Western blot analysis. The results showed that these proteins were down-regulated by miR-200b mimics and up-regulated by miR-200b inhibitors (Fig. 1c, d; Additional file 4: Fig. S8c, d), which was consistent with the results of the proteomics assays.
Validation of expression levels of PPP1R2, MARCKS, and GLIPR2 identified by label free (PC3 cell line). a Expression of miR-200b by miR-200b mimics and inhibitor transfection for 48 h (QPCR). b Expression of target proteins by miR-200b mimics and inhibitor transfection for 48 h (QPCR). c Protein level of PPP1R2, MARCKS, and GLIPR2 regulated by miR-200b mimics or inhibitor transfection for 48 h (western blot). d Density ratio of PPP1R2, MARCKS, and GLIPR2/GAPDH on Western blot. *P < 0.05
Potential functions of DEPs by informatics analysis
Specifically, a broadly adopted web-based functional analysis and GO analysis were used to derive biological meaning from proteomics data. The top regulated biological process (BP) and molecular functions (MF) of DEPs are showed according to their ranking in Fig. 2. The top regulated pathways of DEPs according to rank are showed in Fig. 3a. A search of the KEGG PATHWAY database showed that YAP1, TAZ, PP1and PP2A identified by our proteomics method were involved in Hippo signaling pathway, and which may play an important role in AIPC (Fig. 3b).
Gene ontology analysis of differentially-expressed proteins including down-regulated proteins in miR-200b mimics/mimics NC and up-regulated proteins in miR-200b inhibitor/inhibitor NC groups with PANTHER. Blue, the top biological process (BP) ranked according to − log (P value), P < 0.05. Red, the top molecular functions (MF) ranked according to − log (P value), P < 0.05
Pathway analysis of differentially-expressed proteins including down-regulated proteins in miR-200b mimics/mimics NC and up-regulated proteins in miR-200b inhibitor/inhibitor NC groups. a The top pathways analyzed with PANTHER ranked according to − log (P value), P < 0.05. b Differentially-expressed proteins of the Hippo signaling pathway in AIPC cells (KEGG)
Identification of miR-200b target genes and the interaction networks associated with AIPC
TM4SF1, YAP1, PPP1R2, MARCKS, RTN4 were predicted as target genes of miR-200b by miRBase, while GLIPR2 and SUCLG1 were not predicted as target genes (Fig. 4).
The interaction networks among these seven proteins were analyzed by STRING. The result showed that all of these seven proteins were connected mainly by YAP1 and Ubiquitin C (UBC) to form a network (Fig. 5). The active interaction sources came from text-mining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. The minimum required interaction score was medium confidence (0.400). String also revealed enrichment of these seven proteins for the top pathways: (1) hippo signaling pathway; (2) citrate cycle (TCA cycle); (3) propanoate metabolism; (4) TGF-beta signaling pathway; (5) carbon metabolism; (6) neurotrophin signaling pathway; and (7) miRNAs in cancer.
Discussion
In our previous study, we found that microRNA, miR-200b inhibited proliferation of AIPC cells, which may be an important target for therapy. However, downstream mechanism of miR-200b has not been identified. miRNAs play their roles by inhibiting their target proteins expression. Proteomics would enable the unbiased comparison of different cellular states in biology and medicine at a systems-wide level. Deep proteomics analyses are necessary to characterize the complete scenario of signaling pathways and biologic processes altered as a result of the specific state of cells or tissues [18]. Thus, proteomics method combined with bioinformatics is an ideal strategy to analyze the exact relationship between miRNA-target proteins interaction and the resulting phenotype.
The proteins regulated by miR-200b were identified and analyzed by PANTHER, and relative specific instead of common annotations were taken into account. The results showed that miR-200b may involve a variety of disparate biological processes and cellular functions, such as drug metabolic process, ferric iron transport, mesenchyme migration, nucleosome assembly, and regulation of protein phosphatase type 2A activity. This polytropism reflects that miRNAs may have multiple targets.
In order to reveal the direct link between miR-200b and the resultant phenotypic changes in AIPC, proteins expressed by target genes of miR-200b should be elucidated. It is remarkable that our label-free quantification approach was found to be highly efficient: seven proteins were identified and five of these were products of target genes predicted by miRBase. There are some proteins which are known to play a role in prostate cancer, but were not identified in this study, such as Bmi-1 [19], PDGF-D [20]. One reason may the false-negative results of our proteomics method. Current mass spectrometry techniques cannot identify all proteins in the whole protein of the sample, especially some low abundance proteins. Therefore, in some experiments, mass spectrometry data of some proteins cannot be collected, which is a common phenomenon. Another important reason may be that these proteins may not be involved in the androgen independence of AIPC. Some of these proteins were reported to be related with AIPC. YAP was recently identified as a novel regulator of prostate cancer cell motility, invasion, and androgen independent growth [21, 22] and was considered as a potential therapeutic target for metastatic AIPC. YAP1 is a transcriptional regulator which can act both as a co-activator and a co-repressor; it is a critical downstream regulatory target in the Hippo signaling pathway that plays a pivotal role in organ size control and tumor suppression by inhibiting proliferation and promoting apoptosis [23]. RTN4 is a developmental neurite growth regulatory factor which was shown to inhibit axon–axon adhesion and growth, facilitate neurite branching, and to be potentially involved in proliferation, apoptosis and invasiveness of cancer cells [24]. A recent study showed that RTN4 was regulated by AR in AIPC; however, its function was not clarified [25]. TM4SF1, originally described as “TAAL6”, is a tumor-associated antigen found in various human epithelial malignancies including breast, ovarian, lung, and colon carcinomas [26, 27]. Recent studies indicated that TM4SF1 plays a critical role in cancer cell migration and invasion [28]; it was also shown to be an androgen-responsive protein [29]. MARCKS is the most prominent cellular substrate for protein kinase C. This protein binds calmodulin, actin, and synapsin. MARCKS is a filamentous (F) actin cross-linking protein [30]. Constitutively phosphorylated MARCKS in PC3 cells could contribute to the increased motility and invasiveness shown by this line compared with LNCaP cells [31]. A previous study suggested that MARCKS partly mediated the positive effect of miR-21 on AIPC cell invasion [32]. It is the first time we have reported the proteins being regulated by miR-200b in AIPC.
YAP1 and RTN4 are involved in Hippo signaling pathway. Hippo signaling pathway is the top pathway enriched by String, which indicates that it may be the main signaling pathway regulated by miR-200b. However, there is no definitive evidence of the interaction and functional relationship between TM4SF1, PPP1R2, MARCKS, C9orf19 and SUCLG1. This indicates that there are several target proteins regulated by miR-200b in AIPC; further, these target proteins perform a wide range of functions and are part of a complex network. This is largely attributable to the inherent characteristics of miRNAs, in that these bind to the 3′UTR of the mRNA target through imperfect complementarity or via multiple sites.
The present study reveals the target proteins, biological process and molecular functions regulated by miR-200b. These proteins may be used as new target for AIPC therapy. Further studies should be focused on the interaction network of miR-200b/target proteins, to coordinate playing a role and be applied in therapy of AIPC.
Conclusion
Using a combination of proteomics and bioinformatics, we identified several target proteins of miR-200b in the context of AIPC, TM4SF1, YAP1, PPP1R2, MARCKS, RTN4, GLIPR2, and SUCLG1. These proteins may contribute to the phenotype resulting from miR-200b and may serve as candidate therapeutic targets in AIPC.
Abbreviations
- AIPC:
-
androgen-independent prostate cancer
- CRPC:
-
castration-resistant prostate cancer
- ADT:
-
androgen deprivation therapy
- PANTHER:
-
protein analysis through evolutionary relationships
- mimics-NC:
-
negative miRNA control
- inhibitor-NC:
-
negative inhibitor control
References
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29.
Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: adaptive responses in the androgen axis. Cancer Treat Rev. 2014;40:426–33.
He M, Liu Y, Deng X, Qi S, Sun X, Liu G, et al. Down-regulation of miR-200b-3p by low p73 contributes to the androgen-independence of prostate cancer cells. Prostate. 2013;73:1048–56.
Xu G, Wu J, Zhou L, Chen B, Sun Z, Zhao F, et al. Characterization of the small RNA transcriptomes of androgen dependent and independent prostate cancer cell line by deep sequencing. PLoS ONE. 2010;5:e15519.
Makarova JA, Shkurnikov MU, Wicklein D, Lange T, Samatov TR, Turchinovich AA, et al. Intracellular and extracellular microRNA: an update on localization and biological role. Prog Histochem Cytochem. 2016;51:33–49.
Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–63.
Chua JH, Armugam A, Jeyaseelan K. MicroRNAs: biogenesis, function and applications. Curr Opin Mol Ther. 2009;11:189–99.
Wang L, Song G, Chang X, Tan W, Pan J, Zhu X, et al. The role of TXNDC5 in castration-resistant prostate cancer-involvement of androgen receptor signaling pathway. Oncogene. 2015;34:4735–45.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(− Delta Delta C(T)) method. Methods. 2001;25:402–8.
Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62.
Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–89.
Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805.
Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–42.
Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73.
Zeng Y, Zhang L, Zhu W, Xu C, He H, Zhou Y, et al. Quantitative proteomics and integrative network analysis identified novel genes and pathways related to osteoporosis. J Proteomics. 2016;142:45–52.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52.
Gamez-Pozo A, Berges-Soria J, Arevalillo JM, Nanni P, Lopez-Vacas R, Navarro H, et al. Combined label-free quantitative proteomics and microRNA expression analysis of breast cancer unravel molecular differences with clinical implications. Cancer Res. 2015;75:2243–53.
Yu J, Lu Y, Cui D, Li E, Zhu Y, Zhao Y, et al. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating Bmi-1. Oncol Rep. 2014;31:910–8.
Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–21.
Zhang L, Yang S, Chen X, Stauffer S, Yu F, Lele SM, et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol Cell Biol. 2015;35:1350–62.
Thoma C. Prostate cancer: targetable YAP1-AR interaction key to disease progression. Nat Rev Urol. 2015;12:596.
Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–9.
Pula B, Werynska B, Olbromski M, Muszczynska-Bernhard B, Chabowski M, Janczak D, et al. Expression of Nogo isoforms and Nogo-B receptor (NgBR) in non-small cell lung carcinomas. Anticancer Res. 2014;34:4059–68.
Cheng Y, Yu P, Duan X, Liu C, Xu S, Chen Y, et al. Genome-wide analysis of androgen receptor binding sites in prostate cancer cells. EXP THER MED. 2015;9:2319–24.
Lin CI, Merley A, Sciuto TE, Li D, Dvorak AM, Melero-Martin JM, et al. TM4SF1: a new vascular therapeutic target in cancer. Angiogenesis. 2014;17:897–907.
Visintin A, Knowlton K, Tyminski E, Lin CI, Zheng X, Marquette K, et al. Novel anti-TM4SF1 antibody-drug conjugates with activity against tumor cells and tumor vasculature. Mol Cancer Ther. 2015;14:1868–76.
Allioli N, Vincent S, Vlaeminck-Guillem V, Decaussin-Petrucci M, Ragage F, Ruffion A, et al. TM4SF1, a novel primary androgen receptor target gene over-expressed in human prostate cancer and involved in cell migration. Prostate. 2011;71:1239–50.
Fajardo AM, MacKenzie DA, Olguin SL, Scariano JK, Rabinowitz I, Thompson TA. Antioxidants abrogate alpha-tocopherylquinone-mediated down-regulation of the androgen receptor in androgen-responsive prostate cancer cells. PLoS ONE. 2016;11:e151525.
Green TD, Crews AL, Park J, Fang S, Adler KB. Regulation of mucin secretion and inflammation in asthma: a role for MARCKS protein? Biochim Biophys Acta. 2011;1810:1110–3.
Lang SH, Hyde C, Reid IN, Hitchcock IS, Hart CA, Bryden AA, et al. Enhanced expression of vimentin in motile prostate cell lines and in poorly differentiated and metastatic prostate carcinoma. Prostate. 2002;52:253–63.
Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–5.
Authors’ contributions
MYH participated in the study design, conducted the experiments and helped in manuscript writing; MZG, MQ and WX performed the experiments; ZCJ and WJW analyzed the data; SCZ and YWL participated in the study design and manuscript writing. All authors read and approved the final manuscript.
Acknowledgements
None.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This study was supported by National Natural Science Foundation of China (No. 81302229), Natural Science Foundation of Guangdong Province (No. 2016A030313583), Science and Technology planning Projects of Guangdong Province (No. 2013B051000050, No. 2014A020212538, No. 2016A020215175), the Medical Scientific Research Foundation of Guangdong Province (No. A2016555), the Outstanding Youths Development Scheme of Nanfang Hospital, Southern Medical University (No. 2015J005), and the Science and Technology planning Project of Guangzhou (No. 201704020070).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1.
Primers sequence used in this study.
Additional file 2.
Proteins changed with miR-200b mimics transfection identified by label free proteomics (ratio > 2, P value < 0.05).
Additional file 3.
Proteins up-regulated by miR-200b inhibitor identified by label free proteomics (ratio > 2, P value < 0.05).
Additional file 4.
Figure S1. MS/MS spectra of TM4SF1. Figure S2. MS/MS spectra of YAP1. Figure S3. MS/MS spectra of PPP1R2. Figure S4. MS/MS spectra of MARCKS. Figure S5. MS/MS spectra of RTN4. Figure S6. MS/MS spectra of GLIPR2. Figure S7. MS/MS spectra of SUCLG1. Figure S8. Expression validating of PPP1R2, MARCKS, and GLIPR2 (LnCap-AI cells). a Expression of miR-200b by miR-200b mimics and inhibitor transfection for 48 h (QPCR). b Expression of target proteins by miR-200b mimics and inhibitor transfection for 48 h (QPCR). c Expression of PPP1R2, MARCKS, and GLIPR2 by miR-200b mimics and inhibitor transfection for 48 h (western blot). d Density ratio of PPP1R2, MARCKS, and GLIPR2 / GAPDH on Western blot. *P < 0.05.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
He, M., Gou, M., Qi, M. et al. Label free quantitative proteomics reveals the role of miR-200b in androgen-independent prostate cancer cells. Clin Proteom 15, 8 (2018). https://doi.org/10.1186/s12014-018-9185-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12014-018-9185-1