Abstract
Background
Cluster headache is a severe headache disorder with unknown aetiology. The pathophysiology and symptoms present certain common features with migraine. Specifically, activation of the trigeminal vascular system seems to be involved in both disorders, which is hypothesized to result in neurogenic inflammation and vasodilation of the cerebral vessels. In addition, genetic factors have been implicated in both migraine and cluster headache.
Objective
In order to determine whether or not migraine and cluster headache share genetic risk factors, we screened two genetic variants known to increase the risk of migraine in Sweden in a Swedish cluster headache case-control study population.
Methods
In all, 541 patients and 581 control subjects were genotyped for rs1835740 in close proximity to MTDH (metadherin) and rs2651899 in the PRDM16 (PR/SET domain 16) gene, using TaqMan® real-time PCR and pyrosequencing. In addition, we analyzed MTDH gene expression in a subset of the material, using reverse transcription real-time PCR to determine relative mRNA levels in primary fibroblast cell lines from patients and controls.
Results
We found a trend for association between rs1835740, which is reported to affect MTDH mRNA levels, and cluster headache in our Swedish case-control material (p = 0.043, Χ2 = 4.102). This association was stronger in a subgroup of patients suffering from both cluster headache and migraine (p = 0.031, Χ2 = 6.964). We could further confirm that rs1835740 has an effect on the transcriptional activity of MTDH. In this Swedish cluster headache cohort we did not find an association with the rs2651899 variant.
Conclusions
We conclude that rs1835740 is a potential risk factor for cluster headache in Sweden. Our data indicates that rs1835740 and MTDH might be involved in neurovascular headaches in general whilst rs2651899 is specifically related to migraine.
Similar content being viewed by others
Background
Migraine and cluster headache (CH) are two primary headache disorders that share pathological features and a majority of CH patients are successfully treated with drugs used for migraine, such as triptans [1,2,3,4]. Both disorders are characterized by activation of the trigeminovascular system and neurogenic inflammation [5]. There are also phenotypic similarities between CH and migraine, such as recurring attacks of headache, lateralized pain and associated autonomic symptoms [3, 4]. However in migraine these symptoms are typically nausea and vomiting while in CH autonomic symptoms are e.g. rhinorrhea, lacrimation and ptosis. Sometimes tearing of the eye and nasal congestion at the site of pain as seen in CH can occur also in migraine. However, there are marked differences in the clinical manifestations and associated symptoms between migraine and CH. In particular, the diurnal and seasonal rhythmicity with which headache attacks can be observed in many CH patients is unparalleled in other primary headaches [6, 7].
Genetic factors are likely to influence the risk of developing both migraine and CH [8]. The heritability of CH is unclear, but having a first- or second-degree relative with CH greatly increases the risk of developing the disorder [9]. One study reports heritability (h2) for CH of 0.26, but this number is probably an underestimation since only 2.3% of the patients in the cohort reported a familial history [10]. In our Swedish CH material, 11.2% of the patients report that they have a first-, second- or third degree relative with CH [11]. Several candidate genes such as HCRTR2 (hypocretin receptor 2), ADH4 (alcohol dehydrogenase 4), NOS (nitric oxide synthase) and CLOCK (Circadian Locomotor Output Cycles Kaput) have been suggested to be associated with the disease, although results are conflicting [12,13,14,15,16,17,18,19]. In contrast the heritability of migraine has been estimated to be around 50% (h2 = 0.44) [8, 20]. The contribution of genetic factors to migraine has further been confirmed in several genome-wide association studies (GWAS) [21,22,23,24,25].
We have previously replicated the finding of an association for two of the established risk alleles for migraine in a Swedish migraine case-control population, rs2651899 in PRDM16 (PR/SET domain 16) and rs1835740 located between MTDH (metadherin) and PGCP (carboxypeptidase Q) on chromosome 8q22.1 [26]. Association of the PRDM16 gene to migraine has been confirmed in several replication studies and meta-analyses and it is now considered a well-established migraine candidate gene [27,28,29,30,31,32]. Replication studies on rs1835740 are less affirmative [33,34,35,36]. The MTDH gene was found to associate with migraine in a gene-based multimarker analysis, but the rs1835740 variant did not reach genome-wide significance in a large GWAS meta-analysis [24, 31].
There are indications of CH being more common in families with migraine [37]. Moreover there are genes that have been suggested as candidate genes for both disorders, for example the MTHFR (methylenetetrahydrofolate reductase), HCRTR2 and iNOS (inducible NOS) genes, but there are no conclusive reports on shared genetic risk factors [13, 16, 38,39,40,41,42,43].
The aim of this study was to investigate whether migraine and CH might share genetic determinants since similar pathophysiological events occur in both disorders. Analysis of the distribution of the genetic migraine markers rs2651899 and rs1853740 has not previously been performed in CH. We therefore screened a large Swedish CH case-control population and performed an association analysis of these two markers that have been shown to increase the risk for migraine in Sweden.
Material and methods
Genetic analysis
The material consisted of 541 CH cases and 581 controls where 571 of the controls were anonymous healthy blood donors, these samples were obtained from local blood donation centres in 2003–2005. DNA was isolated from whole blood according to standard procedures after written informed consent. Participating research subjects were required to fill in a questionnaire assessing clinical and lifestyle parameters. Medical journals of all 541 CH patients were reviewed by a neurologist (one of the co-authors A.S., E.W. or C.S.) in order to confirm the diagnosis according to the International Classification of Headache Disorders (ICHD-II) [44]. 518 CH patients were from our carefully characterized Swedish CH biobank recruited during the years of 2013–2017 [11]. 23 of the CH cases were obtained from the TwinGene study, conducted between 2004 and 2008, the procedure of identifying individuals with cluster headache, blood sampling, genotyping and quality control of the array data has been described in previous publications [26, 45]. In total, we analyzed 581 control individuals (56.3% male) and 541 unrelated CH cases with typical CH characteristics; predominantly male (67.7%), mean age of onset was 31.5 years. 464 patients (85.8%) suffered from episodic CH and 50 patients (9.2%) suffered from chronic CH at the time of sample collection, while 27 patients had un uncertain clinical phenotype. 10.9% had familial history of CH defined as having at least one first- or second degree relative with CH (a total of 467 patients provided information on heredity) and 16.4% also suffered from migraine (a total of 451 patients provided information on migraine status). All experiments were approved by the Regional Ethical Review Board, Stockholm, Sweden.
Genotyping was done with quantitative real-time PCR (qPCR), using an ABI 7500 Fast system (Applied Biosystems, Foster City, CA, USA) and TaqMan® reagents for allelic discrimination. TaqMan® genotyping master mix and predesigned single nucleotide polymorphism (SNP) assays for the two SNPs (assay IDs C_11334245_10 (rs1835740) and C_27103073_10 (rs2651899)) were ordered from Applied Biosystems/Life Technologies (Life Technologies Europe BV, Stockholm, Sweden). 2-5 ng dried DNA was used for each reaction, water served as negative control. The cycler was programmed as follows: pre-PCR read 60 °C for 1 minute and enzyme activation at 95 °C for 10 min, 50 cycles of 95 °C for 15 s and 60 °C for 1 minute, and post-PCR read 60 °C for 1 minute. Allelic discrimination was determined using the 7500 software v2.3. The call rate was 98.9% for C_11334245_10 and 98.6% for C_27103073_10.
Distinction between rs1835740 genotypes with the TaqMan® assay C_11334245_10 needed to be confirmed with an additional technique due to weak signal. One randomly chosen 96-whole plate of samples including all three potential genotypes was therefore re-genotyped using pyrosequencing. Primers were designed using ApE v2.0.49 (http://biologylabs.utah.edu/jorgensen/wayned/ape/), verified with the mfold software v3.6 and NCBI online Blast tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and ordered at Thermo Fisher Scientific (Thermo Fisher Scientific, Hägersten, Sweden) [46]. Forward primer (5’-GTTGGAAGTGGGTGTCAGACC-3′), biotinylated reverse primer (5’-GCAGACTTTGGACAGTTCAGAA-3′), and sequencing primer (5’-AACTTGATTCCAATC-3′). DNA fragments were amplified with a regular PCR. The amplified fragments were then fixed to streptavidin-sepharose beads using a PyroMark Vacuum Prep tool (Biotage AB, Uppsala, Sweden), washed and released into an annealing plate containing the pyrosequencing primer. This plate was incubated at 80 °C for 2 min to anneal the primer followed by processing in an automated pyrosequencing PSQ 96 System using PyroMark Gold reagents (QIAGEN Nordic, Sollentuna, Sweden). The results, which were analyzed with the software provided with the PSQ 96 System and manually reviewed, were found to correspond to those obtained with TaqMan® qPCR.
Gene expression analysis
Primary fibroblast cell lines derived from 12 CH patients and 12 control subjects were obtained from skin biopsies performed on the inside of research subjects’ upper arm. The biopsies were obtained under local anaesthesia, after informed consent. All experiments were approved by the Regional Ethical Review Board, Stockholm, Sweden. The biopsy was dissected into several smaller pieces and immediately cultured under a coverslip according to a protocol described by Akira Takashima [47]. Once established, the fibroblast cell lines were passaged two times, frozen in fetal bovine serum (FBS) with 10% DMSO and kept at − 150 °C for long-term storage.
Cells from the 12 CH patients and 12 control subjects were thawed and cultured in 75-cm2 culture flasks (Corning Inc., Corning, NY, USA) using DMEM GlutaMAX™ medium (25 mM D-Glucose and 1 mM sodium pyruvate) supplemented with 15% FBS, 1% 10 mM HEPES and 1% Antibiotic-Antimycotic (10,000 U/mL penicillin, 10,000 mg/mL streptomycin, and 25 mg/mL Amphotericin B), (all from Invitrogen, Paisley, UK), in a humidified 37 °C, 5% CO2 incubator. Cells were passaged once, and then harvested according to a protocol described by Johansson et al. [48]. RNA was prepared directly from frozen cell pellets using an RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Further, the QuantiTect Reverse Transcription Kit (QIAGEN) was used to prepare cDNA from a total of 2 μg RNA.
The reverse transcription qPCR (qRT-PCR) was performed using a standard qRT-PCR program with 45 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 minute. We used an ABI 7500 Fast instrument (Applied Biosystems) and a master mix consisting of Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific), primers at a concentration of 0.25 μM and approximately 200 ng cDNA. Primer sequences for MTDH and for the commonly used reference gene Pyruvate Dehydrogenase E1 Beta (PDHB) were obtained from the literature and ordered from Thermo Fisher Scientific [49, 50]. Both primer pairs had similar R2 values and efficiency: MTDH 0.996/ 105.9% and PDHB 0.990/ 101.8%. One control sample was a suspected outlier and was removed from the analysis after verification with Grubb’s test (Z > 2.4, significance level 0.05). A sensitivity analysis including all samples was performed, which gave a slightly lower p-value and thereby not affecting the outcome of the analysis. MTDH mRNA levels were normalized to the expression of PDHB and further normalized to a reference sample consisting of pooled cDNA from all 24 individuals.
Statistical analysis
Genotype and allele association was analyzed using a two-tailed chi-squared (Χ2) test in GraphPad Prism v5.04 (GraphPad Softwares Inc., La Jolla, CA, USA). Our study design also comprised a subgroup analysis, stratifying patients according to migraine status to evaluate if the occurrence of migraine might affect the results. We did not perform power calculations prior to the study since our sample size was limited by sample availability. A post hoc power calculation based on the available sample size and a minor allele frequency (MAF) of 0.2 indicated that we can detect an odds ratio (OR) < 0.637 or > 1.485 with 80% power, which is within the same order of magnitude as our results (ORrs1835740 = 1.25, 95% confidence interval (CI) 1.01–1.53). Both SNPs were tested for Hardy-Weinberg equilibrium (HWE) in both patients and controls using the online OEGE HWE calculator [51].
The 7500 software v2.3 (Applied Biosystems), GraphPad Prism v5.04 and GraphPad QuickCalcs online tools (GraphPad Softwares Inc) were used for gene expression data analysis. mRNA levels were compared using an unpaired two-tailed t-test. Grubb’s test was used to discern outliers.
Results
We genotyped 541 CH patients and 581 controls for the rs2651899 and rs1835740 SNPs, and tested for association with CH; results are represented in Table 1. For rs1835740, we found a weak association between the rare allele (T) and an increased risk for CH (Chi-square (Χ2) =4.1, OR 1.25, 95% CI 1.01–1.53, p-value = 0.043). Both the TT and the CT genotypes were more common in patients than controls, but there was no significant genotypic association. There was no association between rs2651899 in PRDM16 and CH in our material (Table 1). Both SNPs were in HWE in patients and controls (data not shown).
Since these two SNPs are known to influence the risk of migraine, and many CH patients in our material also suffer from migraine, we performed a stratified analysis with respect to migraine. Patients were separated into two groups; one consisting of patients suffering from CH only, and one consisting of CH patients with migraine. The allelic association of rs1835740 became stronger when analyzing CH patients with migraine, CH patients without migraine, and controls (Χ2 = 6.96, p-value = 0.031) (Table 2). When comparing the allele and genotype frequencies, the mutated allele became increasingly more common in the group of patients suffering from both CH and migraine; 21.1% in CH patients and 26.2% in CH patients with migraine, as compared to the control group where the MAF was 18.5%. The stratified analysis did not affect the results for rs2651899.
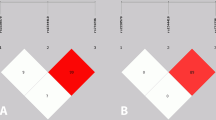
The genetic association discovered here between CH and rs1835740, in combination with the discovery study reporting that rs1835740 potentially affects the expression levels of the MTDH gene lead us to also investigate the MTDH mRNA expression levels in a subset of patients and controls [21]. mRNA was obtained from primary fibroblast cell lines provided from 12 patients and 12 control individuals and analyzed using qRT-PCR. MTDH mRNA expression was normalized to PDHB, which was used as a reference gene, and then to a reference sample. One control individual was identified as an outlier using Grubb’s test and was removed from the analysis. We found a significant difference in the mRNA levels of MTDH when comparing individuals carrying two C alleles (wild-type) at rs1835740, with individuals having one or two T alleles, p = 0.0318, Fig. 1a. There was only one individual carrying the minor allele at both positions in our material, which is the reason we could not compare the three individual genotypes. We further analyzed the mRNA levels with respect to CH diagnosis, as can be observed in Fig. 1b, control subjects and CH patients had similar MTDH mRNA levels (p = 0.195).
MTDH mRNA expression is affected by rs1835740. MTDH mRNA expression levels were normalized to the reference gene PDHB and to a reference sample consisting of pooled cDNA from all individuals. Horizontal bars represent group means, SEM in error bars, groups were compared using an unpaired, two-tailed t-test. a) mRNA levels are compared between individuals with two major alleles (CC) and individuals carrying one or more T alleles (CT + TT) at rs1835740 regardless of headache diagnosis, p = 0.0318. b) mRNA levels were compared between healthy control subjects and individuals with cluster headache, p = 0.195
Discussion and conclusion
We have analyzed a Swedish CH population for two SNPs that have previously been identified as genetic risk factors for migraine in Sweden and globally [21, 22]. One of the variants, rs1835740, was found to be associated with an increased risk for CH. The other SNP, rs2651899, was not associated with CH in Sweden.
A more detailed analysis of rs1835740 revealed that the association was even stronger in a subgroup of patients that have both CH and migraine. When comparing the MAF between the three groups, we found that the minor allele (T) was present in 26.2% of the individuals suffering from both disorders, which is 7.7% more than among controls, a significant difference (p = 0.016) with an OR of 1.56 (95% CI 1.10–2.21), and 5.1% more than in CH without migraine. This difference was not significant possibly because of the smaller sample sizes in the stratified analysis. Our findings thus appear to indicate that the T allele is enriched in the patient group having both CH and migraine. In a former GWAS suggesting MTDH as a candidate gene for migraine, the effect of the rs1835740 association was stronger in a subgroup of patients suffering from migraine with aura (MA) than in patients with migraine without aura (MO), which is also an indication of this SNP being associated with more complex headache phenotypes [21]. Interestingly, in a study on rs1835740 and migraine by Azimova et al. (2015) small groups of CH patients (n = 9) and chronic tension type headache patients (n = 20) were used as control groups. In their study, all chronic tension headache patients were wild type for rs1835740, while the MAF of CH patients was 44.4% [36]. One possible limitation of our study is the lack of clinical information related to our control material. We cannot exclude the occurrence of headache in the control group; statistically, the occurrence of CH should be extremely low, or non-existent, but we have to assume that around 15% of our controls also suffer from migraine, corresponding to the prevalence of migraine in the Swedish population. In view of our data, the occurrence of migraine in the control group would imply a higher MAF of rs1835740, and should therefore not introduce a risk of false positive results.
MTDH has been suggested to be a likely candidate gene for migraine because of its proximity to the intergenic SNP rs1835740. Moreover, rs1835740 was found to affect the gene expression of MTDH, thus providing an appealing pathophysiological mechanism for migraine: MTDH is known to downregulate EAAT2 (excitatory amino acid transporter 2), a major glutamate transporter, and could thus affect glutamate regulation at a synaptic level [52, 53]. We studied the MTDH mRNA levels in individuals with different rs1835740 genotype, and also analyzed expression with respect to CH diagnosis. Similarly to previous reports, the relative quantification showed a difference in expression between individuals with different genotypes for rs1835740 [21]. Individuals carrying one or two T alleles had significantly higher mRNA levels than individuals with the CC genotype. eQTLs (expression quantitative trait loci) may vary between different types of tissue and although gene expression in primary fibroblasts does not reveal how MTDH is expressed in nervous tissue, we were encouraged to be able to confirm the initial finding that associated the T allele of rs1835740 with increased MTDH expression in immortalized lymphocytes [21, 54]. There was no difference in mRNA levels between CH patients and controls.
The discovery of rs1835740 and MTDH provided a molecular link between migraine and familial hemiplegic migraine (FHM), a familial form of migraine caused by mutations in ion channels, since mutations associated with FHM subtypes one and two are known to cause increased levels of glutamate at the synaptic cleft [55, 56]. Similarly, our results suggest that MTDH and glutamatergic mechanisms may be involved also in CH pathophysiology. There are previous indications that glutamate might play a role in CH as well. A recent publication demonstrated a decrease in kynurenine metabolites, known glutamate receptor antagonists, in CH patients [57]. This could potentially affect the activity of the glutamate receptor NMDA and nociceptive sensitization [57, 58].
We conclude that rs1835740 in MTDH is associated not only with migraine but also with CH, whilst rs2651899 in PRDM16 seems to be specifically related to migraine in Sweden. In view of our stratified analysis showing that the T allele of rs1835740 was more common in patients with both disorders, we also suggest that rs1835740 might constitute a genetic risk factor for more complex headache phenotypes, for example a combination of different primary headache syndromes as illustrated in our study. The T allele of rs1835740 was further shown to affect MTDH mRNA expression in primary fibroblasts. In depth analyses of MTDH and the rs1835740 SNP in CH and in other primary headache disorders would be of interest in order to determine to what extent rs1835740 is related to severe headache.
Abbreviations
- ADH4 :
-
Alcohol dehydrogenase 4
- CH:
-
Cluster headache
- CI:
-
Confidence interval
- CLOCK :
-
Circadian Locomotor Output Cycles Kaput
- DF:
-
Degrees of freedom
- eQTL:
-
Expression quantitative trait loci
- FBS:
-
Fetal bovine serum
- FHM:
-
Familial hemiplegic migraine
- GWAS:
-
Genome-wide association study
- HCRTR2 :
-
Hypocretin receptor 2
- HWE:
-
Hardy-Weinberg equilibrium
- ICHD:
-
International Classification of Headache Disorders
- iNOS :
-
Inducible NOS
- MA:
-
Migraine with aura
- MAF:
-
Minor allele frequency
- MO:
-
Migraine without aura
- MTDH :
-
Metadherin
- MTHFR :
-
Methylenetetrahydrofolate reductase
- NOS :
-
Nitric oxide synthase
- OR:
-
Odds ratio
- PDHB :
-
Pyruvate Dehydrogenase E1 Beta
- PGCP :
-
Carboxypeptidase Q
- PRDM16 :
-
PR/SET domain 16
- qPCR:
-
Quantitative real-time PCR
- qRT-PCR:
-
Reverse transcription qPCR
- SNP:
-
Single nucleotide polymorphism
- χ2 :
-
Chi-squared
References
Bahra A, May A, Goadsby PJ (2002) Cluster headache: a prospective clinical study with diagnostic implications. Neurology 58:354–361
Robbins MS, Starling AJ, Pringsheim TM, Becker WJ, Schwedt TJ (2016) Treatment of cluster headache: the American headache society evidence-based guidelines. Headache J Head Face Pain. 56:1093–1106
The International Headache Society (IHS) (2018) Headache classification Committee of the International Headache Society (IHS) the international classification of headache disorders, 3rd edition. Cephalalgia 38:1–211
Vollesen AL, Benemei S, Cortese F et al (2018) Migraine and cluster headache – the common link. J Headache Pain 19:89
Buture A, Gooriah R, Nimeri R, Ahmed F (2016) Current understanding on pain mechanism in migraine and cluster headache. Anesthesiol pain Med 6:e35190
Kudrow L (1987) The cyclic relationship of natural illumination to cluster period frequency. Cephalalgia 7(Suppl 6):76–78
Ofte HK, Berg DH, Bekkelund SI, Alstadhaug KB (2013) Insomnia and periodicity of headache in an Arctic cluster headache population. Headache J Head Face Pain. 53:1602–1612
Mulder EJ, Van Baal C, Gaist D et al (2003) Genetic and environmental influences on migraine: a twin study across six countries. Twin Res 6:422–431
Bjørn Russell M (2004) Epidemiology and genetics of cluster headache. Lancet Neurol 3:279–283
Montagna P, Mochi M, Prologo G et al (1998) Heritability of cluster headache. Eur J Neurol 5:343–345
Steinberg A, Fourier C, Ran C, Waldenlind E, Sjöstrand C, Belin AC (2018) Cluster headache - clinical pattern and a new severity scale in a Swedish cohort. Cephalalgia 38:1286–1295
Rainero I, Gallone S, Valfrè W et al (2004) A polymorphism of the hypocretin receptor 2 gene is associated with cluster headache. Neurology 63:1286–1288
Weller CM, Wilbrink LA, Houwing-Duistermaat JJ et al (2015) Cluster headache and the hypocretin receptor 2 reconsidered: a genetic association study and meta-analysis. Cephalalgia 35:741–747
Rainero I, Rubino E, Gallone S et al (2010) Cluster headache is associated with the alcohol dehydrogenase 4 (ADH4) gene. Headache 50:92–98
Fourier C, Ran C, Steinberg A, Sjöstrand C, Waldenlind E, Carmine Belin A (2016) Screening of two ADH4 variations in a Swedish cluster headache case-control material. Headache 56:835–840
Sjöstrand C, Modin H, Masterman T, Ekbom K, Waldenlind E, Hillert J (2002) Analysis of nitric oxide synthase genes in cluster headache. Cephalalgia 22:758–764
Rainero I, Rivoiro C, Gallone S et al (2005) Lack of association between the 3092 T→C clock gene polymorphism and cluster headache. Cephalalgia 25:1078–1081
Cevoli S, Mochi M, Pierangeli G et al (2008) Investigation of the T3111C CLOCK gene polymorphism in cluster headache. J Neurol 255:299–300
Fourier C, Ran C, Zinnegger M et al (2018) A genetic CLOCK variant associated with cluster headache causing increased mRNA levels. Cephalalgia 38:496–502
Svensson DA (2004) Etiology of primary headaches: the importance of genes and environment. Expert Rev Neurother 4:415–424
Anttila V, Stefansson H, Kallela M et al (2010) Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet 42:869–873
Chasman DI, Schurks M, Anttila V et al (2011) Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat Genet 43:695–698
Freilinger T, Anttila V, de Vries B et al (2012) Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat Genet 44:777–782
Ligthart L, de Vries B, Smith AV et al (2011) Meta-analysis of genome-wide association for migraine in six population-based European cohorts. Eur J Hum Genet 19:901–907
Cox HC, Lea RA, Bellis C et al (2012) A genome-wide analysis of “bounty” descendants implicates several novel variants in migraine susceptibility. Neurogenetics 13:261–266
Ran C, Graae L, Magnusson PKE, Pedersen NL, Olson L, Belin AC (2014) A replication study of GWAS findings in migraine identifies association in a Swedish case-control sample. BMC Med Genet 15:38
Sintas C, Fernández-Morales J, Vila-Pueyo M et al (2015) Replication study of previous migraine genome-wide association study findings in a Spanish sample of migraine with aura. Cephalalgia 35:776–782
Fan X, Wang J, Fan W et al (2014) Replication of migraine GWAS susceptibility loci in Chinese Han population. Headache 54:709–715
An XK, Ma QL, Lin Q, Zhang XR, Lu CX, Qu HL (2013) PRDM16 rs2651899 variant is a risk factor for chinese common migraine patients. Headache 53:1595–1601
Anttila V, Winsvold BS, Gormley P et al (2013) Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet 45:912–917
Gormley P, Anttila V, Winsvold BS et al (2015) Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. bioRxiv 48:16–18
Esserlind A-L, Christensen AF, Le H et al (2013) Replication and meta-analysis of common variants identifies a genome-wide significant locus in migraine. Eur J Neurol 20:765–772
Sintas C, Carreno O, Fernandez-Morales J et al (2012) A replication study of a GWAS finding in migraine does not identify association in a Spanish case-control sample. Cephalalgia 32:1076–1080
Ghosh J, Pradhan S, Mittal B (2013) Genome-wide-associated variants in migraine susceptibility: a replication study from North India. Headache 53:1683–1694
Stovner L, Hagen K, Jensen R et al (2007) The global burden of headache: a documentation of headache prevalence and disability worldwide. Cephalalgia 27:193–210
Azimova J, Kondratieva N, Sergeev A et al (2015) The role of polymorphism of regulatory region of MTDH gene (Rs1835740) in migraine and other forms of primary headaches. J Neurol Stroke 3:101
Kudrow L, Kudrow DB (1994) Inheritance of cluster headache and its possible link to migraine. Headache J Head Face Pain. 34:400–407
Schürks M, Neumann FA, Kessler C et al (2011) MTHFR 677C>T Polymorphism and Cluster Headache. Headache J Head Face Pain 51:201–207
Samaan Z, Gaysina D, Cohen-Woods S et al (2011) Methylenetetrahydrofolate reductase gene variant (MTHFR C677T) and migraine: a case control study and meta-analysis. BMC Neurol 11:66
Rainero I, Rubino E, Valfrè W et al (2007) Association between the G1246A polymorphism of the hypocretin receptor 2 gene and cluster headache: a meta-analysis. J Headache Pain. 8:152–156
Pinessi L, Binello E, De Martino P et al (2007) The 1246G→A polymorphism of the HCRTR2 gene is not associated with migraine. Cephalalgia 27:945–949
De OS, Mansur T, Gonçalves FM, Martins-Oliveira A et al (2012) Inducible nitric oxide synthase haplotype associated with migraine and aura. Mol Cell Biochem 364:303–308
Gonçalves FM, Luizon MR, Speciali JG, Martins-Oliveira A, Dach F, Tanus-Santos JE (2012) Interaction among nitric oxide (NO)-related genes in migraine susceptibility. Mol Cell Biochem 370:183–189
The International Headache Society (IHS) (2004) The international classification of headache disorders: 2nd edition. Cephalalgia 24 Suppl1:9–160
Ekbom K, Svensson DA, Pedersen NL, Waldenlind E (2006) Lifetime prevalence and concordance risk of cluster headache in the Swedish twin population. Neurology 67:798–803
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Takashima A (1998) Establishment of fibroblast cultures. Curr Protoc Cell Biol 2:1–12
Johansson A-S, Owe-Larsson B, Hetta J, Lundkvist GB (2016) Altered circadian clock gene expression in patients with schizophrenia. Schizophr Res 174:17–23
Wang Y, Wei Y, Tong H et al (2015) MiR-302c-3p suppresses invasion and proliferation of glioma cells via down-regulating metadherin (MTDH) expression. Cancer Biol Ther 16:1308–1315
Häbig K, Gellhaar S, Heim B et al (2013) LRRK2 guides the actin cytoskeleton at growth cones together with ARHGEF7 and tropomyosin 4. Biochim Biophys Acta - Mol Basis Dis 1832:2352–2367
Rodriguez S, Gaunt TR, Day INM (2008) Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol 169:505–514
Kang D-C, Su Z-Z, Sarkar D, Emdad L, Volsky DJ, Fisher PB (2005) Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene 353:8–15
Lee S-G, Kim K, Kegelman TP et al (2011) Oncogene AEG-1 promotes glioma-induced neurodegeneration by increasing glutamate excitotoxicity. Cancer Res 71:6514–6523
Bullaughey K, Chavarria CI, Coop G, Gilad Y (2009) Expression quantitative trait loci detected in cell lines are often present in primary tissues. Hum Mol Genet 18:4296–4303
Anttila V, Wessman M, Kallela M, Palotie A (2011) Towards an understanding of genetic predisposition to migraine. Genome Med 3:17
Pietrobon D (2007) Familial hemiplegic migraine. Neurotherapeutics 4:274–284
Curto M, Lionetto L, Negro A et al (2015) Altered serum levels of kynurenine metabolites in patients affected by cluster headache. J Headache Pain. 17:27
Vécsei L, Szalárdy L, Fülöp F, Toldi J Kynurenines in the CNS: recent advances and new questions. Nat. Rev. Drug Discov.. Nature Publishing Group 2013, 12:64–82
Acknowledgements
We thank Nancy Pedersen for contributing with data from the TwinGene study and Ann-Christin Karlsson for organizing the recruitment of patients.
Funding
This work was financed by Lennanders stiftelse, the Swedish Brain Foundation, Swedish Research Foundation, and Karolinska Institutet Research funds.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon request.
Author information
Authors and Affiliations
Contributions
CR performed genotyping and gene expression experiments, the statistical analysis and drafted the manuscript. CF and MZ prepared DNA, performed genotyping and revised the manuscript critically for important intellectual content. AS, CS, EW recruited and verified the clinical diagnosis of all patients, contributed to the scientific content and revised the manuscript critically. ACB conceived and planned the study, was involved in data analysis, drafting and revising the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Clinical information and biological samples were collected from the study participants after written informed consent. The experiments described herein were approved by the Regional Ethical Review Board in Stockholm, Sweden.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ran, C., Fourier, C., Zinnegger, M. et al. Implications for the migraine SNP rs1835740 in a Swedish cluster headache population. J Headache Pain 19, 100 (2018). https://doi.org/10.1186/s10194-018-0937-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s10194-018-0937-0