Abstract
Background
HDAC4 is a key regulator of chondrocyte hypertrophy and skeletal development, but it is not clear whether the increase in vascular invasion at growth plates is related to HDAC4 expression. To determine it, we investigated the relationship between HDAC4 and angiogenesis in both in vivo and in vitro models.
Methods
HDAC4 was deleted in Col2α1-Cre; HDAC4fl/fl mice. Growth of the Col2α1-Cre; HDAC4d/d mice was compared with HDAC4fl/fl mice at postnatal days 2, 4, 6, and 8. X-rays were taken to examine skeletal development. At postnatal days 14 and 21, mice were euthanized for specimen collection. Murine chondrocytes were isolated from the ventral parts of rib cages of 6-day-old mice (C57Bl/6) and transfected with a vector expressing HDAC4 as a fusion protein with green fluorescent protein (GFP). Relative expression levels of HDAC4, VEGF, and Hif1α were measured in these cells by Western blot, RT-qPCR, enzyme-linked immunosorbent, histology, and immunohistochemistry assays.
Results
The Col2α1-Cre; HDAC4d/d mice were markedly smaller compared with the control mice. At postnatal days 14 and 21, the Col2α1-Cre; HDAC4d/d mice exhibited a shortened growth plate, a larger secondary ossification center, and stronger staining of CD31 and CD34 compared to control mice. The isolated chondrocyte cells exhibited a high transfection efficiency of HDAC4 which resulted in the detection of a significant decrease in VEGF and Hif1α levels compared with the control chondrocytes.
Conclusions
HDAC4 expression in chondrocytes contributes to angiogenesis in the growth plate, and its absence in vivo negatively affects growth plates.
Similar content being viewed by others
Background
During endochondral ossification, mesenchymal cells differentiate into chondrocytes which eventually form a cartilage growth plate. Ossification begins when hypertrophic chondrocytes undergo programmed cell death and the resulting calcified cartilage is invaded by blood vessels, osteoblasts, osteoclasts, and mesenchymal precursors to form primary ossification centers. Within these centers, bone eventually replaces the disappearing cartilage. Degradation and remodeling of the cartilage matrix are essential for vascular invasion and angiogenesis, and this step is considered to be a crucial step in endochondral ossification. This process has been shown to be coordinately controlled by several signaling molecules, including HDAC4, fibroblast growth factor 2 (FGF-2), bone morphogenetic protein 2 (BMP2), vascular endothelial growth factor (VEGF), and Indian hedgehog (IHH) proteins (Chen et al. 2016b; Huang et al. 2016a, b).
Histone deacetylase 4 (HDAC4) is a key member of a family of tissue-specific class II HDAC proteins. HDAC4 is expressed in prehypertrophic chondrocytes in the growth plate and is a negative regulator of growth plate maturation by inhibiting matrix metalloproteinase (MMP) -13 expression and inhibiting the activity of runt-related transcription factor-2 (Runx2) which is essential for chondrocyte hypertrophy during endochondral bone formation (Vega et al. 2004; Li et al. 2014; Cao et al. 2014). We previously observed that expression of HDAC4 significantly decreased in hypertrophic chondrocytes present in the growth plates of newborn mice (Guan et al. 2011). While it is well established that HDAC4 is a key regulator of chondrocyte hypertrophy and skeletal development (Wang et al. 2014a, b), it is not clear whether the increase in vascular invasion at the hypertrophic zone and bone junction area is related to HDAC4 expression.
Other scholars investigated the relationship between HDAC4 regulation of HIF-1, VEGF and angiogenesis in cancer cell line (Geng et al. 2011; Ellis et al. 2009). Geng H et al. found that HDAC4 inhibition led to the reduction of HIF-1-mediated target gene expressions and activities in cancer cells under hypoxic condition. However HDAC4 inhibition had little effect on Hif1α under normal condition. And they suggested that not all HIF-1 target genes were down-regulated by HDAC4 inhibition (Geng et al. 2011). Ellis L et al. reviewed the influence of HIF-1α and VEGF on tumor angiogenesis and how HDACs played a critical role in HIF-1α transcriptional activity. The studies presented that HDAC4 played an important role in hypoxic induced angiogenesis, and that pharmalogical inhibition and shRNA against HDAC4 offered a new strategy in anti-cancer therapy through decreasing HIF-1α expression and inhibiting angiogenesis in tumor (Ellis et al. 2009). However the relationship of HDAC4 regulation of HIF-1, VEGF and vascular invasion at the hypertrophic zone and bone junction area remains unclear.
To examine the significance of chondrocyte-derived HDAC4 for angiogenesis at the hypertrophic zone and bone junction area, we investigated the relationship between HDAC4 and angiogenesis in both in vivo and in vitro models. Since global deletion of HDAC4 is lethal in mice (Vega et al. 2004), we generated a conditional knockout mouse model of HDAC4 to perform in vivo studies. For an in vitro model, HDAC4 was overexpressed as a fusion protein with green fluorescent protein (GFP) in chondrocytes. In these models, the role of HDAC4 in relation to angiogenesis and angiogenesis-related proteins was examined, respectively.
Methods
Generation of Floxed HDAC4 animals
Approval for the animal experiments conducted in this study was obtained from the Institutional Animal Care and Use Committee at Rhode Island Hospital. Col2α1-Cre mice were mated to HDAC4fl/fl (from Dr. Olson, University of Texas Southwestern Medical Center) animals to obtain HDAC4fl/−, Col2α1-Cre mice (Vega et al. 2004). Mice transgenic for Cre in collagen type 2α1-expressing chondrocytes (Col2α1-Cre) have been previously reported (Long et al. 2001). These HDAC4fl/−, Col2α1-Cre mice were subsequently interbred with HDAC4fl/fl animals. Their offspring (Col2α1-Cre; HDAC4d/d) and their littermates (HDAC4fl/fl) were analyzed. Their littermates, HDAC4fl/fl animals, were used as the control. Routine mouse genotyping was performed with the following forward and reverse primers for the Cre allele and the HDAC4 allele: 5′-ATCCGAAAAGAAAACGTTGA-3′ and 5′-ATCCAGGTTACGGATATAGT-3′, and 5′-ATCTGCCCACCAGAGTATGTG-3′ and 5′-CTTGTTGAGAACAAACTCCTGCAGCT-3′, respectively in each case. The expected product sizes were: 620 bp for the Cre allele, 480 bp for the wild-type HDAC4 allele, and 480 bp and 620 bp for the floxed HDAC4 allele.
We divided the experiment into two groups (HDAC4fl/fl control group and Col2α1-Cre; HDAC4d/d group). There were 8 mice in each group. Mice were euthanized at postnatal days 14 and 21 for specimen collection.
Radiography
Mice were anesthetized by 25 mg/kg xylazine and 75 mg/kg ketamine. X-rays were taken with Faxitron MX-20 Cabinet X-ray (Faxitron, Arizona, USA) and used to evaluate skeletal development.
Cell culture and transfections
Chondrocyte cells were isolated from the ventral parts of rib cages that were resected from 6-day-old mice (C57Bl/6). The cells were subsequently cultured in F-12 media supplemented with 10% fetal bovine serum (FBS; Gibco BRL, USA) at 37 °C in 5% CO2 as previously described (Chen et al. 2016a). For transfections, the cells were plated in 6-well plates and grown overnight. When the density of the monolayer reached 70–80% confluency, 1 ml of medium containing serum and antibiotics was added to each well 30–60 min before transfection. Meanwhile, 0.5 μg, 1 μg, 1.5 μg, and 2.0 μg of a vector containing GFP fused to HDAC4 DNA were each combined with 100 μl of serum-free, high glucose DMEM. The preparations were vortexed gently to mix them. In separate tubes, 3.0 μl of GenJetTM reagent (Ver. II) (SignaGen Laboratories, Ijamsville, MD, USA) was added into 100 ul aliquots of serum-free, high glucose DMEM. The latter preparations were added to the former preparations, with gently pipetting performed to mix them. After a 15 min incubation at room temperature, the GenJetTM-DNA complexes were gently added drop-wise into individual wells and then the plates were swirled to provide homogeneous application of the transfection-DNA complexes onto the cells. The transfected cells were then cultured in a humidified incubator under 5% CO2 and hypoxia (2% O2) (NuAire Autoflow NU8500 Water Jacket CO2 incubator, Plymouth, MN, USA). Forty-eight hours later, the percentage of positively transfected cells (e.g., those expressing GFP) was determined for each sample with a fluorescence microscope (E800; Nikon, Tokyo, Japan). Approximately 300 cells from three independent experiments were scored for each sample.
Western blot analysis
Forty-eight hours after the chondrocytes were transfected with a vector expressing HDAC4, they were washed with ice-cold PBS and lysed in RIPA buffer (50 mM Tris·HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 1% NP-40) at 4 °C. After 30 min, the lysates were cleared by centrifugation for 20 min at 4 °C. Total protein in each sample was quantified with a BCA Protein Assay Reagent Kit (Pierce, Rockford, IL, USA). Western blotting was performed according to standard procedures. Briefly, the proteins were electrophoresed in 10% SDS-PAGE gels and then transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with anti-HDAC4 (sc-46,672, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-actin (Cell Signaling Technology, Danvers, MA, USA), anti-VEGF (Santa Cruz, CA, USA), and anti-Hif1α (Cell Signaling Technology, Danvers, MA, USA) antibodies, with each at a concentration of 0.2 μg/ml. The membranes were subsequently incubated with peroxidase-conjugated mouse anti-goat (sc-2354, Santa Cruz, CA, USA) and goat anti-mouse (sc-2005, Santa Cruz, CA, USA) secondary antibodies (diluted 1:1000) as appropriate. The relative intensities of HDAC4, VEGF, and Hif1α expression were semi-quantitated by densitometry and normalized to levels of β-actin expression by using Image J software (U.S. National Institutes of Health, Bethesda, MD, USA), as previously described (Li et al. 2014).
Real-time quantitative PCR (qPCR)
RNA was isolated from the chondrocytes, which were transfected with 2.0 μg GFP-HDAC4 for forty-eight hours, with RNAqueous kit (Ambion, Austin, TX). After treatment with TURBO DNase (Ambion), 1 μg of RNA was reverse-transcribed with random hexamers to obtain first-strand cDNA using iScript cDNA kit (Bio-Rad). The quantification of mRNA for Hif1α and VEGF was performed by two-step real time quantitative RT-PCR (Qiagen, Valencia, CA) (n = 3). Simultaneously, controls were conducted in the absence of HDAC4. Primer pairs were as following: for Hif1α, ACA-GTG-GTA-CTC-ACA-GTC-GG (forward) and CCC-TGC-AGT-AGG-TTT-CTG-CT (reverse), for VEGF, ACC-AGC-GCA-GCT-ATT-GCC-GT (forward) and CAC-CGC-CTT-GGC-TTG-TCA-CA (reverse), for 18S, CGG-CTA-CCA-CAT-CCA-AGG-AA (forward) and GCT-GGAATT-ACC-GCG-GCT (reverse). Relative transcript levels were calculated according to the equation x = 2-△△Ct, where △△Ct = △Ct E - △Ct C, △Ct E = Ctexp - Ct18S, and △Ct C=Ct C-Ct 18S) (Fang et al. 2001; Wang et al. 2014a).
Enzyme-linked Immunosorbent assay (ELISA)
Aliquots of medium from each transfected well were collected and levels of VEGF were measured with a VEGF Immunoassay kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions (Sun et al. 2009).
Histology
Right knee joints were harvested from animals which were euthanized on postnatal days 14 and 21. The knee joints were fixed in 4% neutral buffered formalin for 24 h at 20 °C and then were processed without decalcification and embedded in a single block. Sections were cut from the sagittal plane, with adjacent sections collected at intervals of 0, 100, and 200 μm from the cartilage layer. Two serial 6-μm-thick sections from each interval were stained with Safranin O and then mounted on slides in order to evaluate development of the growth plate and secondary ossification center. Thickness of the growth plate and ratio of the area of secondary ossification center to the area of tibial plateau were subsequently quantitated from images collected with a Nikon Ri 1 microscope (Nikon, Melville, NY, USA).
Immunohistochemistry
Briefly, 6-μm sections of knee joint tissue were placed on positively charged glass slides (Thermo Fisher Scientific, Asheville, NC, USA) and then dried with a hotplate to increase the adherence of the tissues to the slides. After the samples were deparaffinized and rehydrated according to conventional methods, endogenous peroxidases were blocked with 3% hydrogen peroxide in methanol (Sigma-Aldrich, Burlington, USA). After 30 min the samples were digested with 5 mg/ml hyaluronidase in phosphate buffered saline (PBS) (Sigma-Aldrich) for 20 min and then were incubated with anti-CD31 (1:100 dilution) (SC-56; Santa Cruz, CA, USA) and anti-CD34 (1:100 dilution) (Santa Cruz, CA, USA) antibodies overnight at 4 °C. Following treatment of the samples with appropriate biotinylated secondary antibodies, a 3,3’diaminobenzidine (DAB) streptavidin-peroxidase (SP) DAB Histostain-SP immunohistochemistry kit (ZYMED203 Laboratories/Invitrogen, Carlsbad, CA, USA) was used to visualize bound antibodies. After the sections were counterstained with hematoxylin (ZYMED Laboratories/Invitrogen), the tissues were imaged with a Nikon Ri 1 microscope (Nikon, Melville, NY, USA) (Guan et al. 2012).
Statistical analysis
GraphPad 6 software (GraphPad, San Diego, CA, USA) was used to perform statistical analyses.
Data are expressed as the mean ± standard error of the mean (SEM). One-way analysis of variance (ANOVA) Dunnett’s multiple comparisons test was used to compare all data associated with the control and GFP-HDAC4 groups. P-values < 0.05 were considered statistically significant.
Results
Generation of Col2α1-Cre; HDAC4d/d mice
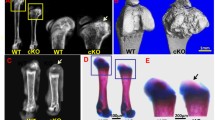
To investigate the function of HDAC4 in vivo, Col2α1-Cre; HDAC4d/d mice were used in this study to delete HDAC4 in chondrocytes. Wild-type, homologous, and heterozygous genotypes were confirmed by Southern blot PCR. Growth of the Col2α1-Cre; HDAC4d/d mice was subsequently compared with wild-type (HDAC4f1/f1) mice at postnatal days 2, 4, 6, and 8, with x-rays taken to examine skeletal development. The Col2α1-Cre; HDAC4d/d mice were markedly smaller compared to the HDAC4f1/f1 mice (the control group) (Fig. 1), indicating that Col2α1-Cre; HDAC4d/d mice undergo slower development when HDAC4 is disrupted.
Col2α1-Cre; HDAC4d/d mice have extremely shortened growth plates and enlarged secondary ossification centers
Safranin O/Fast green staining was performed on paraffin sections of knee joints resected from Col2α1-Cre; HDAC4d/d and HDAC4f1/f1 mice at postnatal days 14 and 21. The Col2α1-Cre; HDAC4d/d mice exhibited a shortened growth plate and larger secondary ossification center compared with the HDAC4f1/f1 mice (Fig. 2a and b). It is noted that the reduction of thickness in three zones (resting, proliferating and hypertrophic) of growth plate was significant for the Col2α1-Cre; HDAC4d/d mice at postnatal days 14. HDAC4 knockout has small effect on the thickness reduction of different zones at postnatal days 21.
Shortened growth plates and larger secondary ossification centers in Col2α1-Cre; HDAC4d/d mice. Safranin O/Fast green staining was performed on paraffin sections at days 14 and 21. a Shortened growth plates and larger secondary ossification centers were observed in the Col2α1-Cre; HDAC4d/d mice compared with the HDAC4f1/f1 mice on postnatal days 14 and 21. b Significant differences in the thicknesses of the growth plates and ratio of area of secondary ossification center to area of tibial plateau were observed between the Col2α1-Cre; HDAC4d/d and HDAC4f1/f1 groups on postnatal days 14 and 21. Data are expressed as mean ± SEM. *P < 0.05 vs. the vehicle group
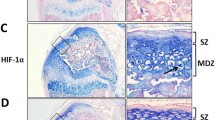
Immunohistochemistry of Col2α1-Cre; HDAC4d/d mice detected stronger CD31 and CD34 expression
Sections of knee joint tissues collected from Col2α1-Cre; HDAC4d/d and HDAC4f1/f1 mice on postnatal days 14 and 21 were subjected to immunohistochemistry assays to detect expression of CD31 and CD34. Both proteins exhibited stronger staining in the Col2α1-Cre; HDAC4d/d sections than in the control sections (Fig. 3), and the stronger staining appeared in the secondary ossification center of Col2α1-Cre; HDAC4d/d mice. These results suggest that vascular invasion is more pronounced when HDAC4 is disrupted in Col2α1-Cre; HDAC4d/d mice.
Increased expression of CD31 and CD34 in Col2α1-Cre; HDAC4d/d mice. Strong staining of CD31 (a) and CD34 (b) was observed in knee joint tissue sections from Col2α1-Cre; HDAC4d/d mice compared with knee joint sections from control animals on postnatal days 14 and 21. The red arrow indicates the newly formed vessels
Overexpression of HDAC4 in vitro
To establish an in vitro model of HDAC4 overexpression, chondrocyte cells were isolated from the ventral parts of rib cages that were resected from 6-day-old mice (C57Bl/6). After these cells were established as in vitro cultures, the cells were transfected with various doses of a vector expressing HDAC4 as a fusion protein with GFP. Transfection efficiency was evaluated 48 h after transfection based on the numbers of chondrocyte cells expressing GFP. The percentages of GFP expressing cells according to GFP-HDAC4 dose were: 84.3% (79–91%) for 0.5 μg, 83.3% (76–88%) for 1.0 μg, 83.3% (78–88%) for 1.5 μg, and 84.6% (80–90%) for 2.0 μg (Fig. 4).
Transfection efficiency of HDAC4 into isolated chondrocytes. a The transfection efficiency of various doses of GFP-HDAC4 DNA into isolated murine chondrocytes was examined with confocal laser scanning microscopy based on GFP expression. Nuclei are stained with Hoechst 33342 (Hoechst). b Approximately 300 cells from three independent experiments were scored. Data are expressed as mean ± SEM
Transfection of GFP-HDAC4 leads to a decrease in levels of VEGF and Hif1α
Western blotting was performed to detect the levels of HDAC4, VEGF, and Hif1α proteins in chondrocyte cells following overexpression of HDAC4 (Fig. 5a, b). Relative expression levels of VEGF exhibited a significant decrease in the chondrocyte cells that were transfected with 1.5 μg GFP-HDAC4 or 2.0 μg GFP-HDAC4 compared with the control group. Relative expression levels of Hif1α were also significantly lower in the chondrocyte cells transfected with 1.0 μg GFP-HDAC4 and 2.0 μg GFP-HDAC4 compared with the control cells.
Levels of HDAC4, VEFG, and Hif1α following HDAC4 overexpression. a, b Expression levels of HDAC4, Hif1α, and VEGF proteins were detected in Western blots following the transfection of various doses of GFP-HDAC4 DNA into isolated murine chondrocytes. Detection of β-actin was performed as a loading control. The accompanying bar graphs present the average relative intensity values for VEGF and Hif1α, respectively, from three independent experiments. Data are expressed as mean ± SEM. *P < 0.05 vs. vehicle group
The effect of HDAC4 overexpression on the mRNA level of VEGF and Hif1α was determined by RT-qPCR (Fig. 6a, b). The results indicated that the relative mRNA levels of VEGF and Hif1α reduced in the chondrocytes transfected with GFP-HDAC4 compared with the control group.
mRNA levels of VEFG and Hif1α following HDAC4 overexpression. a, b mRNA levels of Hif1α and VEGF were detected in RT-qPCR following the transfection of 2.0 μg GFP-HDAC4 into isolated murine chondrocytes. The accompanying bar graphs present the average relative mRNA values for VEGF and Hif1α, respectively, from three independent experiments. Data are expressed as mean ± SEM. *P < 0.05 vs. vehicle group
Overexpression of HDAC4 leads to a decrease in VEGF level in the media
Forty-eight hours after transfection with GFP-HDAC4, samples of media were collected to detect levels of VEGF. The ELISA data indicated that increased expression of HDAC4 led to a decrease in the amount of VEGF into the media (Fig. 7). Moreover, the difference in the amount of secreted VEGF compared with the control group was significant for the 1.0 μg GFP-HDAC4, 1.5 μg GFP-HDAC4, and 2.0 μg GFP-HDAC4 groups.
Discussion
During skeletal development, the appendicular skeleton and part of the axial skeleton are formed largely by endochondral ossification. The latter is a sequential process that involves the proliferation, differentiation, cell death, and resorption of chondrocytes. Skeletal vasculature plays a significant role in the process of bone development (including endochondral and intramembranous ossification), as well as in the regeneration and remodeling of bone (Dai and Rabie 2007; Hankenson et al. 2011; Huang et al. 2016a). Blood vessels supply bones with needed nutrients, including oxygen, hormones, and growth factors, thereby regulating the process of bone development and remodeling (Filipowska et al. 2017). Secretion of VEGF by chondrocytes is also essential for the coupling of osteogenesis and angiogenesis (Hu and Olsen 2016). HDAC4 has been shown to be a critical negative regulator of chondrocyte hypertrophy based on its ability to bind and inhibit Runx2, a transcription factor that is critical for chondrocyte hypertrophy (Zhou et al. 2015) and whose expression promotes both bone metastasis and osteolysis (Trotter et al. 2015). To date, there is no direct evidence that expression of HDAC4 decreases blood vessel formation in the growth plate, although it has been demonstrated that global deletion of HDAC4 is lethal in mice (Vega et al. 2004). To examine the significance of chondrocyte-derived HDAC4 in angiogenesis, we selectively ablated the HDAC4 gene from collagen type II-expressing cells by using Cre/loxP gene targeting. As a result, severely runted and slow-growing mice were generated. Moreover, compared with the HDAC4f1/f1 group, shortened growth plates were observed with Safranin O/Fast green staining and a larger ratio of ossification center to tibia was measured for the Col2α1-Cre; HDAC4d/d mice by postnatal days 14 and 21. The similar result was also found from Col2-Cre; HDAC4f1/f1 mice at birth by Nishimori et al., and however the Col2-Cre; HDAC4f1/f1 mice died around days 14 (Nishimori et al. 2019). Increased expression of CD31 and CD34 was detected in the knee joints of the Col2α1-Cre; HDAC4d/d mice by postnatal days 14 and 21 as shown in Fig. 3a and b. Both CD31 and CD34 are widely used as markers of angiogenesis, hematopoietic cells, and endothelial progenitor cells (Hassanein et al. 2017). Thus, the present data are consistent with a role for HDAC4 in both angiogenesis and osteoblast differentiation via regulation of Runx2 and osteocalcin expression (Sun and Beier 2014; Lawson et al. 2013).
VEGF is an important mediator of angiogenesis (Dai and Rabie 2007), while Hif1α is a transcription factor that stimulates the growth of new blood vessels during angiogenesis (Filipowska et al. 2017). It has been shown that vascular endothelial cells that are localized near the chondro-osseous junction express higher levels of Hif1α and exhibit strong expression of CD31 (Kusumbe et al. 2014). When HDAC4 was overexpressed in isolated murine chondrocytes, a significant decrease in VEGF and Hif1α protein expression was detected by Western blot. The HDAC4 overexpression also repressed the mRNA level of VEGF and Hif1α as determined by RT-qPCR. In addition, a significant decrease in the secretion of VEGF was further detected by ELISAs. However, this was inconsistent with the previous reports, in which Ablating HDAC4 signaling induced Hif1α protein acetylation, reduced Hif1α protein level in cancer cell lines (Geng et al. 2011; Isaacs et al. 2013; Ellis et al. 2009), and inhibited endothelial tumor angiogenesis (Geng et al. 2011; Cadot et al. 2009; Liu et al. 2009; Mottet et al. 2009). One of the reasons may be related to the cell type. In the previous reports, the relationship of HDAC4 and Hif1α was investigated in cancer cell lines or for tumor angiogenesis. In the present study, the chondrocytes that were transfected with higher doses of HDAC4 vector exhibited lower levels of VEGF and Hif1α expression compared with the cells that were transfected with lower doses of HDAC4 vector. The present results are consistent with our CD31 and CD34 straining results in which specific deletion of HDAC4 from chondrocytes in vivo led to a strong staining of CD31 and CD34. These results verify that HDAC4 is a critical regulator of angiogenesis and development of cartilage growth plates, and they support our hypothesis that overexpression of HDAC4 may inhibit angiogenesis in growth plates by decreasing levels of VEGF and Hif1α. However, it is noted that VEGF/ Hif1α is not the only factor promoting angiogenesis. There may be other unknown ways involving the results due to deletion of HDAC4. Gene array is going to be conducted to explore the unknown pathway in the future study.
Conclusions
In the present study, selective gene targeting was achieved to specifically ablate the HDAC4 gene from collagen type 2α1-expressing cells. Following this ablation, the affected mice exhibited a remarkable phenotype which included a shorter growth rate, enhanced angiogenesis, the formation of premature bone, and an enlarged secondary ossification center. Correspondingly, our in vitro results also demonstrated a role for HDAC4 in angiogenesis, and they identified VEGF and Hif1α as downstream targets.
Availability of data and materials
All relevant data supporting the conclusions of this article is included within the manuscript.
Abbreviations
- BMP2:
-
Bone morphogenetic protein 2
- ELISA:
-
Enzyme-linked immunosorbent assay
- FGF-2:
-
Fibroblast growth factor 2
- GFP:
-
Green fluorescent protein
- HDAC4:
-
Histone deacetylase 4
- Hif1α:
-
Hypoxia-inducible factor 1α
- IHH:
-
Indian hedgehog
- MMP-13:
-
Matrix metalloproteinase 13
- Runx2:
-
Runt-related transcription factor-2
- VEGF:
-
Vascular endothelial growth factor
References
Cadot B, Brunetti M, Coppari S, Fedeli S, de Rinaldis E, Dello Russo C, et al. Loss of histone deacetylase 4 causes segregation defects during mitosis of p53-deficient human tumor cells. Cancer Res. 2009;69:6074–82.
Cao K, Wei L, Zhang ZQ, Guo L, Zhang CM, Li YP, Sun CQ, Sun XJ, Wang SW, Li PC, Wei XC. Decreased histone deacetylase 4 is associated with human osteoarthritis cartilage degeneration by releasing histone deacetylase 4 inhibition of runt-related transcription factor-2 and increasing osteoarthritis-related genes: a novel mechanism of human osteoarthritis cartilage degeneration. Arthritis Res Ther. 2014;16(6):491.
Chen CW, Wei XC, Wang SW, Jiao Q, Zhang Y, Du GQ, Wang XH, Wei FY, Zhang JZ, Wei L. Compression regulates gene expression of chondrocytes through HDAC4 nuclear relocation via PP2A-dependent HDAC4 dephosphorylation. Biochim Biophys Acta. 2016a;1863(7 Pt A):1633–42.
Chen W, Sheng PY, Huang ZY, Meng FG, Kang Y, Huang GX, Zhang ZQ, Liao WM, Zhang ZJ. MicroRNA-381 regulates chondrocyte hypertrophy by inhibiting histone Deacetylase 4 expression. Int J Mol Sci. 2016b;17(9):1377–88.
Dai J, Rabie AB. VEGF: an essential mediator of both angiogenesis and endochondral ossification. J Dental Res. 2007;86(10):937–50.
Ellis L, Hammers H, Pili R. Targeting tumor angiogenesis with histone deacetylase inhibitors. Cancer Lett. 2009;280:145–53.
Fang J, Yan L, Shing Y, Moses MA. HIF-1alpha-mediated up-regulation of vascular endothelial growth factor, independent of basic fibroblast growth factor, is important in the switch to the angiogenic phenotype during early tumorigenesis. Cancer Res. 2001;61(15):5731–5.
Filipowska J, Tomaszewski KA, Niedå wiedzki Å, Walocha JA, Niedå wiedzki T. The role of vasculature in bone development, regeneration and proper systemic functioning. Angiogenesis. 2017;20(3):291–302.
Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue CH, Qian DZ. HDAC4 protein regulates HIF1 protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286(44):38095–102.
Guan Y, Chen Q, Yang X, Haines P, Pei M, Terek R, Wei XC, Zhao TC, Wei L. Subcellular relocation of histone deacetylase 4 regulates growth plate chondrocyte differentiation through Ca2+/calmodulin-dependent kinase IV. Am J Physiol Cell Physiol. 2012;303(1):33–40.
Guan YJ, Yang X, Wei L, Chen Q. MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J. 2011;25(12):4457–66.
Hankenson KD, Dishowitz M, Gray C, Schenker M. Angiogenesis in bone regeneration. Injury. 2011;42(6):556–61.
Hassanein SM, Sahar MA, Hassanein MHNE, Hanaa AA, Adel EA, Moustafa EH, Abir I. Human umbilical cord blood CD34-positive cells as predictors of the incidence and short-term outcome of neonatal hypoxic-ischemic encephalopathy: a pilot study. J Clin Neurol. 2017;13(1):84–90.
Hu K, Olsen BR. Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair. J Clin Invest. 2016;126(2):509–26.
Huang B, Wang W, Li Q, Wang Z, Yan B, Zhang Z, Wang L, Huang M, Jia C, Lu J, Liu S, Chen H, Li M, Cai D, Jiang Y, Jin D, Bai X. Osteoblasts secrete Cxcl9 to regulate angiogenesis in bone. Nat Commun. 2016a;7:13885.
Huang HM, Li XL, Tu SQ, Chen XF, Lu CC, Jiang LH. Effects of roughly focused extracorporeal shock waves therapy on the expressions of bone morphogenetic protein-2 and osteoprotegerin in osteoporotic fracture in rats. Chin Med J. 2016b;129(21):2567–75.
Isaacs JT, Antony L, Dalrymple SL, Brennen WN, Gerber S, Hammers H, Wissing M, Kachhap S, Luo J, Xing L, Bjork P, Olsson A, Bjork A, Leanderson T. Tasquinimod is an allosteric modulator of HDAC4 survival signaling within the compromised cancer microenvironment. Cancer Res. 2013;73(4):1386–99.
Kusumbe AP, Ramasamy SK, Adams RH. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 2014;507(7492):323–8.
Lawson KA, Teteak CJ, Zou J, Hacquebord J, Ghatan A, Zielinska-Kwiatkowska A, Fernandes RJ, Chansky HA, Yang L. Mesenchyme-specific knockout of ESET histone methyltransferase causes ectopic hypertrophy and terminal differentiation of articular chondrocytes. J Biol Chem. 2013;288(45):32119–25.
Li P, Wei X, Guan Y, Chen Q, Zhao T, Sun C, Wei L. MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development. FASEB J. 2014;28(9):3930–41.
Liu R, Wang L, Chen G, Katoh H, Chen C, Liu Y, et al. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/ histone deacetylatase 4 association to the locus. Cancer Res. 2009;69:2252–9.
Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–108.
Mottet D, Pirotte S, Lamour V, Hagedorn M, Javerzat S, Bikfalvi A, et al. HDAC4 represses p21WAF1/Cip1 expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene. 2009;28:243–56.
Nishimori S, Lai F, Shiraishi M, Kobayashi T, Kozhemyakina E, Yao TP, Lassar AB, Kronenberg HM. PTHrP targets HDAC4 and HDAC5 to repress chondrocyte hypertrophy. JCI Insight. 2019;4(5):1–17.
Sun MM, Beier F. Chondrocyte hypertrophy in skeletal development, growth, and disease. Birth Defects Res C Embryo Today. 2014;102(1):74–82.
Sun X, Wei L, Chen Q, Terek RM. HDAC4 represses vascular endothelial growth factor expression in chondrosarcoma by modulating RUNX2 activity. J Biol Chem. 2009;284(33):21881–90.
Trotter TN, Li M, Pan QY, Peker D, Rowan PD, Li J, Zhan FH, Suva LJ, Javed A, Yang Y. Myeloma cell-derived Runx2 promotes myeloma progression in bone. Blood. 2015;125(23):3598–608.
Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119(4):555–66.
Wang SW, Wei XC, Zhou JM, Zhang J, Li K, Chen Q, Terek R, Fleming BC, Goldring MB, Ehrlich MG, Zhang G, Wei L. Identification of alpha2-macroglobulin as a master inhibitor of cartilage-degrading factors that attenuates the progression of posttraumatic osteoarthritis. Arthritis Rheumatol. 2014a;66(7):1843–53.
Wang ZK, Qin GJ, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics. 2014b;6(1):139–50.
Zhou J, Li PC, Chen Q, Wei XC, Zhao T, Wang ZK, Wei L. Mitogen-activated protein kinase p38 induces HDAC4 degradation in hypertrophic chondrocytes. Biochim Biophys Acta. 2015;1853(2):370–6.
Acknowledgements
None.
Funding
The project was supported by Grant R01AR059142 from NIH/NIAMS. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health and NSFC.
Author information
Authors and Affiliations
Contributions
Study design: L.L.G. and L.W. Study conduct: L.L.G., S.C.L. and G.Q.D. Data collection: L.L.G., S.C.L., and G.Q.D. Data analysis: L.L.G., S.C.L., G.Q.D., D.W., X.C.W. and L.W. Data interpretation: L.L.G., S.C.L., G.Q.D., D.W., X.C.W. and L.W. Drafting manuscript: L.L.G., S.C.L. and L.W. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal experiments were approved by the Institutional Animal Care and Use Committee at Rhode Island Hospital, and all surgical and experimental procedures were performed in accordance with the regulations and guidelines of the Institutional Animal Care and Use Committee at Rhode Island Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gao, L., Li, S., Wei, X. et al. Conditional deletion of HDAC4 from collagen type 2α1-expressing cells increases angiogenesis in vivo. Mol Med 26, 36 (2020). https://doi.org/10.1186/s10020-020-00154-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s10020-020-00154-6