Abstract
Background
The Chinese medicine formulation ISF-1 (also known as Bu-Yang-Huan-Wu-Tang) for post-stroke rehabilitation could increase the expression of growth-regulating protein follistatin by approximately 4-fold. This study aims to identify the active compounds of ISF-1 for the induction of follistatin expression.
Methods
Active compounds in ISF-1 responsible for induction of follistatin were identified by a bioactivity-guided fractionation procedure involving liquid-liquid extraction, HPLC separation and RT-PCR detection. The aqueous extracts of seven ISF-1 ingredients including Semen Persicae (Tao Ren) and the S. Persicae-derived fractions were assayed for the induction of follistatin mRNA expression in human hepatocarcinoma HepG2 cells by RT-PCR. The concentrations of isolated compounds were proportionally normalized to the reported IC50 concentration (5.8 mg/mL) of the formulation ISF-1 in HepG2. The active fractions were characterized by reverse-phase HPLC on a C18 column and identified by mass spectrometry.
Results
Three ingredients of ISF-1, namely S. Persicae (Tao Ren), Pheretima (Di Long), and Flos Carthami (Hong Hua), induced the expression of follistatin mRNA. Among these, the ingredient S. Persicae were the most active, and amygdalin from S. Persicae extract was identified as a novel follistatin inducer. Amygdalin stimulated the growth of skeletal muscle cell line C2C12 cells in a concentration-dependent manner.
Conclusions
Amygdalin isolated from S. Persicae extract in ISF-1 through a bioactivity-guided fractionation procedure induced the expression of follistatin in HepG2 and C2C12 cell lines.
Similar content being viewed by others
Background
Stroke causes severe disability and mortality worldwide, characterized by dramatic structural and metabolic changes in skeletal muscle that can lead to post-stroke sarcopenia [1, 2]. Recombinant tissue plasminogen activator is the only FDA approved drug for the treatment of acute ischemic stroke but is limited by a narrow therapeutic window and the ineffectiveness for post-stroke recovery [3, 4].
Myostatin has emerged as a molecular target for therapeutic intervention [5]. Both pharmacological inhibition and genetic targeting of myostatin were under evaluation for treating sarcopenia [6–9]. Follistatin is an autocrine glycoprotein with strong ability to antagonize different members of the TGF-β superfamily, such as activin and myostatin [10–13], regulating cell proliferation, cell differentiation, inflammatory responses, and embryogenesis [11, 14–16]. Follistatin has been recently evaluated as a treatment for fibrotic diseases in animal models aa [14, 17, 18]. However, as an endogenous antagonist of myostatin, follistatin acts as a negative regulator of muscle growth [13]. Transfection of follistatin cDNA construct for increasing follistatin protein expression could increase muscle mass in experimental animals [6, 7, 19]. Therefore, pharmacological induction of follistatin expression might serve as a new strategy for the treatment of post-stroke and age-related sarcopenia.
The Chinese medicine formulation ISF-1 (also known as Bu-Yang-Huan-Wu-Tang), was used for post-stroke rehabilitation. The formulation consists of seven ingredients, including Radix Astragali (Huang Qi), Radix Angelicae Sinensis (Dang Gui), Radix Paeoniae Rubra (Chi Shao), Rhizoma Chuanxiong (Chuan Xiong), Flos Carthami (Hong Hua), Semen Persicae (Tao Ren), and Pheretima (Di Long) [20, 21]. We previously profiled the cellular responses to formulation ISF-1 in human HepG2 cells by a genome-wide biological response fingerprinting (BioReF) approach [20], and developed a bioactivity-guided fractionation procedure involving liquid-liquid extraction, HPLC separation and RT-PCR detection to identify the active compounds involved in the induction of heme oxygenase-1 and leukotriene B4 12-hydroxydehydrogenase [22, 23].
This study aims to identify the active compounds of ISF-1 for the induction of follistatin expression.
Methods
Cell culture and reagents
The human hepatocellular carcinoma cell line HepG2 [ATCC No. HB-8065] and mouse skeletal muscle cell line C2C12 [ATCC No. CRL-1722] were obtained from American Type Culture Collection (USA). The cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin (Invitrogen, USA) at 37°C in a humidified atmosphere containing 5% CO2 and 95% air. All chemicals were obtained from Sigma-Aldrich Co. (USA), unless otherwise indicated. Dried herbal extracts, R. Astragali (Cat No. 1008; Batch No. A090611-01), S. Persicae (Cat No. 1106; Batch No. A06273-01), Pheretima (Cat No. 1118, Batch No. A06314-01), F. Carthami (Cat No. 1297, Batch No. A04579-02), R. Sinensis (Cat No. 1187, Batch No. A00397-03), R. Rubra (Cat No. 1059, Batch No. A06001-01), and R. Chuanxiong (Cat No. 1019; A06430-01), were purchased from Nong’s Pharmaceutical Ltd. (Hong Kong). Oligonucleotide primers specific for follistatin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were purchased from the Centre for Genomic Sciences, The University of Hong Kong.
Apparatus and chromatographic conditions
HPLC analysis was performed on an Alltima™ HP C18 column (250 × 4.6 mm, 5 μm, Alltech, USA) under the control of a HPLC system (Waters, USA) equipped with a Waters 996 Model photodiode array detector (DAD), a Waters 600S Model system controller, and a gradient generator. The column was run in a gradient mobile phase generated by mixing an aqueous solution (A) and acetonitrile (B). The samples were eluted with 5% B for 5 min, followed by a linear gradient up to 95% B over a period of 50 min. The flow rate was set at 1 mL/min and the column temperature was maintained at 25°C. For analysis of the fifth fraction (fraction F5) and the spiked-in amygdalin standard, the samples were eluted with aqueous solution A containing 9% acetonitrile and 9% methanol. The UV absorbance was recorded over the range of 200–400 nm.
Assay of herbal extracts for induction of follistatin
Dried powder samples of aqueous herbal extracts (R. Astragali, S. Persicae, Pheretima, F. Carthami, R. Sinensis, R. Rubra, and R. Chuanxiong) were purchased from Nong’s Pharmaceutical Ltd. (Hong Kong). For herbal extract preparation, the dried powder of each herbal extract (100 mg) was dissolved in 1 mL of Milli Q water produced by Milli Q Synthesis A10 Water Purificaiton System (EMD Millipore, Germany) at 80°C for 30 min, with vortexing every 10 min. After cooling to room temperature, the insoluble materials were removed by centrifugation at 12,000 rpm on an Eppendorf 5424 microcentrifuge (Eppendorf AG, Germany) for 10 min. The supernatant was recovered and sterilized by passage through a syringe filter with a 0.22-μm (Pall, New York, USA) membrane. For assays of follistatin induction, HepG2 cells were treated for 24 h with the supernatants of the individual herbal extracts at the following concentrations: R. Astragali, 5.18 mg/mL; S. Persicae, 0.13 mg/mL; Pheretima, 0.13 mg/mL; F. Carthami, 0.13 mg/mL; R. Sinensis, 0.26 mg/mL; R. Rubra, 0.19 mg/mL; R. Chuanxiong, 0.13 mg/mL. The expression of follistatin was detected by RT-PCR and analyzed by 1% agarose gel electrophoresis.
Bioactivity-guided fractionation of Semen Persicae extract
The dried S. Persicae extract (50 g) was resuspended in 250 mL of Milli Q water and heated at 80°C for 1 h. Following precipitation with 70% ethanol overnight and removal of insoluble materials by centrifugation at 4000 × g for 15 minutes, the supernatant (A) was collected and sequentially extracted three times with 250 mL of ethyl acetate (EA fraction) and n-butanol (n-butanol fraction). The solvents for each extraction were finally removed by a rotary evaporator (Tokyo Rikakikai, Japan) under vacuum. The dried residues from each preparation were dissolved in dimethyl sulfoxide, filtered through a 0.22-μm membrane (Pall, USA), and subjected to the follistatin induction assay in HepG2 cells.
For isolation of the active compounds, the n-butanol fraction was further separated by semi-preparative reverse-phase HPLC on an Alltima C-18 column (250 × 10.0 mm, 5 μm, Alltech, USA). The column was run in a gradient mobile phase generated by mixing an aqueous solution (A) and acetonitrile (B). The samples were eluted with 5% B for 5 min, followed by a linear gradient elution up to 95% B over a period of 50 min. The flow rate was set at 3 mL/min and the column temperature was maintained at 25°C. The UV absorbance was monitored on a HPLC UV detector (Waters, USA) over the wavelength range of 200–400 nm. Eleven fractions were collected and assayed for follistatin induction. Finally, each active compound was purified as a single peak in the HPLC profile.
Chemical identification by mass spectrometry
The HPLC fraction containing the active compound was analyzed on a Varian MicroSorb C18 column (2 × 150 mm) (Agilent, USA) at a flow rate of 0.8 mL/min under the control of a LC-MS system (Waters, USA) equipped with a 1525 Separations Module and a Waters 2998 DAD Unit (Waters, USA). The mobile phase compositions were water with 0.3% (v/v) formic acid (A) and acetonitrile (B). The gradient was set as follows: 0–8 min, 5- 50% B; 8–10 min, 50% B; 12–15 min, 95% B. The flow rate was constant at 0.8 mL/min and the column temperature was maintained at 20°C. The injection volume was 20 μL. The eluents were analyzed on a triple quadrupole mass spectrometer 3200 QTRAP® system (ABI/Sciex, USA) equipped with an ESI-Turbo V™ source operating in the positive ionization mode under the control of an Analyst v1.4.2 data system (Applied Biosystems/MDS Sciex, Canada). The mass spectrometry analyses were performed under the following conditions: N2 drying gas, 10 L/min; capillary voltage, 20 V; nebulizer pressure, 30 psi; ion spray voltage, 4 kV; and capillary temperature, 325°C.
RT-PCR detection
Following treatment with herbal extracts or isolated fractions, total RNA was isolated from the cells by TRIzol reagent (Invitrogen, USA). The mRNAs were converted into cDNAs by Moloney Murine Leukemia Virus (M-MuLV) reverse transcriptase kit (Fermentas, USA). Human and mouse follistatin mRNAs were detected by PCR using specific oligonucleotide primers as follows: human follistatin mRNA (NM_006350): sense, 5'-GTTTTCTGTCCAGGCAGCTCCACA-3', antisense, 5'-GCAAGATCCGGAGTGCTTTACTTCCA-3'; mouse follistatin mRNA (NM_008046): sense, 5′-CTCTTCAAGTGGATGATTTTC-3', antisense, 5′-ACAGTAGGCATTATTGGTCTG-3′. As an internal control, GAPDH mRNA (NM_001256799) was also detected using specific oligonucleotide primers: sense, 5'-CAAGGTCATCCATGACAACTTTG-3', antisense, 5'-GTCCACCACCCTGTTGCTGTAG-3'. The PCR amplifications were performed as follows: denaturation at 94°C for 3 min; 30 cycles of amplification at 94°C for 30 s, 65°C (human follistatin), 50°C (mouse follistatin), or 55°C (GAPDH) for 30 s, and 72°C for 30 s; and extension at 72°C for 10 min. The PCR products were analyzed by electrophoresis in a 1.0% agarose gel containing Gel Red and visualized under UV light by a GelDoc imaging system (Bio-Rad, USA). The intensity of EtBr-stained gel bands was obtained by densitometric analysis using software Quantity One (Bio-Rad, USA). After normalization against housekeeping gene GAPDH, the mean values of follistatin induction relative to the untreated control were calculated based on the results of three independent experiments.
Cell viability assay
Cell viability was evaluated by a standard colorimetric assay as previously described [24]. Briefly, C2C12 cells (0.4 × 104 cells/100 μL) were seeded in a 96-well microplate and treated with amygdalin at concentrations of 0–25 μM for 24 or 48 h. At the end of the treatment, the cell monolayers were incubated with 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) solution (0.5 mg/mL) in phosphate-buffered saline for 4 h. The formation of purple formazan was measured in relation to the absorbance at 570 nm by a microplate reader (Bio-Rad, USA). The cell viability was presented as a percentage relative to that of vehicle-treated control cells.
Statistical analysis
The results were presented as means ± SD from three independent experiments. Statistical analysis was performed by two-tailed unpaired Student’s t-test with SPSS 13.0 software (SPSS, Chicago, USA). A P value < 0.05 was considered as statistically significant.
Results
Screening of herbal extracts for induction of follistatin expression
We treated HepG2 cells with aqueous extracts of the individual herbs for 24 h. Follistatin induction was subsequently detected by RT-PCR using specific primers. Three herbal extracts (S. Persica e, Pheretima, and F. Carthami) induced the expression of follistatin mRNA, while the expression of the housekeeping gene GAPDH was not affected (Figure 1). The S. Persicae extract showed the strongest activity for increasing follistatin mRNA expression among the three extracts. Thus, the S. Persicae extract was selected for further characterization by bioactivity-guided fractionation.
Identification of the active ingredients for induction of follistatin expression. HepG2 cells were treated with different herbal extracts, R. Astragali, R. Chuanxiong, F. Carthami, Pheretima, R. Sinensis, R. Rubra, and S. Persicae, for 24 h. Subsequently, the expression of follistatin mRNA was detected by RT-PCR and 1 % agarose gel electrophoresis of the PCR products. +, treatment with selected herbal extract; -, no treatment with selected herbal extract.
Bioactivity-guided fractionation of S. Persicae extract for follistatin induction
We developed a simple bioactivity-guided fractionation procedure to isolate the active compounds from S. Persicae extract (Figure 2). The dried aqueous herbal extract purchased from Nong’s Pharmaceutical Ltd. and was verified to have activity in follistatin induction. Briefly, HepG2 cells were treated with S. Persicae extract at contractions of 0, 0.125, 0.25, 0.5, 0.75, and 1.0 mg/mL. Subsequent RT-PCR analyses of follistatin mRNA confirmed S. Persicae extract induced the expression of follistatin mRNA in a concentration-dependent manner (Figure 3A). For bioactivity-guided fractionation, the dried S. Persicae extract was re-extracted and then precipitated with ethanol. The resulting mixture was separated by centrifugation into two parts, a supernatant (S/N) and a pellet. The supernatant was sequentially extracted with ethyl acetate (EA) and n-butanol, giving rise to another three fractions, EA extract, n-butanol extract, and H2O phase. All of the fractions were evaluated for induction of follistatin mRNA. The n-butanol extract effectively induced the expression of follistatin mRNA (Figure 3A).
Schematic illustration of the bioactivity-guided fractionation procedure for identification of the active compound from S. Persicae extract. The n-butanol extract was separated by semi-preparative RP-HPLC on a C18 column. All of the fractions were analyzed by semi-quantitative RT-PCR for follistatin mRNA induction in HepG2 cells.
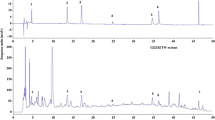
RT-PCR assay for induction of follistatin expression. (A) Assays of S. Persicae extract and liquid-liquid extraction products. HepG2 cells were treated with S. Persicae extract at the indicated concentrations and S. Persicae-derived fractions for 24 h, and the expression of follistatin mRNA was detected by RT-PCR. S/N, supernatant; EA, ethyl acetate. (B) HPLC separation of the n-butanol extract. The n-butanol extract was separated by semi-preparative RP-HPLC on a C18 column into 11 fractions. (C) Assays of HPLC fractions. The HPLC fractions were assayed for induction of follistatin mRNA expression.
Amygdalin in the n-butanol extract of S. Persicae is a novel inducer of follistatin
We separated the n-butanol extraction into 11 fractions by HPLC on a C18 column to isolate the active compound from the n-butanol extract of S. Persicae (Figure 3B). All of the fractions were assessed for induction of follistatin mRNA. Fraction F5 showed the strongest activity in inducing follistatin mRNA expression, while fractions F2 and F9 also showed some activity (Figure 3C). Fraction F5 was further analyzed by LC-MS. The positive ESI-MS m/z spectrum of fraction F5 contained four important signals at 458 [M + H+], 475 [M], 480 [M + Na+], and 937 [2 M + Na+] (Figure 4A), suggesting the presence of amygdalin in this fraction [25].
Chemical characterization of fraction F5. (A) LC-MS analysis of fraction F5. Positive ESI-MS m/z signals are shown. The chemical structure of amygdalin was generated by ACD/Chem Sketch software (http://www.acdlabs.com/resources/freeware/chemsketch/). (B) HPLC verification of amygdalin as the active compound. The fraction F5, amygdalin standard, and F5-amygdalin mixture were sequentially analyzed by reverse-phase HPLC on a C18 column under the same conditions. (C) RT-PCR verification of commercial amygdalin for induction of follistatin mRNA. HepG2 cells were treated with commercial amygdalin at the indicated concentrations for 24 h. The expression of follistatin mRNA was detected by RT-PCR.
We compared the retention times of fraction F5 and an amygdalin standard in the HPLC profile to verify the effect of amygdalin on follistatin mRNA expression. Fraction F5 was confirmed to elute with the same pattern as the commercial amygdalin standard. We observed two closely eluted peaks for both fraction F5 and the amygdalin standard (Figure 4B). Next, we clarified whether the commercial amygdalin standard could induce follistatin mRNA. As expected, the commercial amygdalin standard effectively induced follistatin mRNA expression in a concentration-dependent manner (Figure 4C). LC-MS analyses of fractions F2 and F9 could not identify any chemical compounds reported by others [26, 27].
Effects of amygdalin on the expression of follistatin and proliferation of skeletal muscle C2C12 cells
We treated the cells of mouse skeletal muscle cell line C2C12 with amygdalin at various concentrations (0–25 μM) to characterize whether amygdalin could induce follistatin mRNA expression in skeletal muscle cells. The expression of follistatin mRNA was detected by RT-PCR using specific primers. Amygdalin effectively induced the expression of follistatin mRNA in a concentration-dependent manner (Figure 5A). We further investigated whether amygdalin could accelerate the growth of skeletal muscle cells. We treated C2C12 cells with amygdalin at the same concentrations (0–25 μM) for 24 or 48 h. The viable cells were determined by a standard MTT assay. Amygdalin was confirmed to stimulate the growth of skeletal muscle cell line C2C12 cells in a concentration- and time-dependent manner (Figure 5B).
Effect of amygdalin on follistatin expression and cell proliferation in skeletal muscle C2C12 cells. (A) Induction of follistatin mRNA by amygdalin. Mouse skeletal muscle C2C12 cells were treated with amygdalin at the indicated concentrations for 24 h. The levels of follistatin mRNA were determined by RT-PCR. (B) Stimulation of skeletal muscle cell growth by amygdalin. C2C12 cells were treated with amygdalin at the indicated concentrations for 24 or 48 h. The viable cells were measured by a standard MTT assay. The values represent means ± SD from three independent experiments. *P < 0.05, **P < 0.01, amygdalin treatment vs. control.
Discussion
The genome-wide BioReF approach not only helps in the selection of candidate genes for complex herbal medicines in a bias-free manner, but also enables rapid identification of the corresponding active compounds [20, 22, 23]. Subsequent bioactivity-guided fractionation could be performed on a relatively small scale to identify the active compounds that were actually responsible for the specific bioactivity of the parent herbal extracts. In the present study, we investigated the active compounds for the induction of follistatin, from a previous BioReF profile [20]. We developed a bioactivity-guided fractionation procedure to identify the active compound from formulation ISF-1 involved in the induction of follistatin expression. We identified three herbs, S. Persicae, Pheretima, and F. Carthami, as the active ingredients in HepG2 cells. Amygdalin was identified from the S. Persicae extract as an inducer of follistatin. Amygdalin existed in two forms, D-amygdalin and neoamygdalin [28, 29]. The two closely eluted peaks in the HPLC profiles of fraction F5 and the amygdalin standard were observed, which might reflect the two forms of amygdalin. However, we cannot rule out the possibility that similar or different active compounds might be present in other active fractions (e.g., fractions F2 and F9).
S. Persicae was an anticoagulant, antiphlogistic, and anodyne in Chinese medicine [30, 31]. However, little was known about the role of S. Persicae in the treatment of post-stroke disorders. Several myostatin inhibitors, including myostatin propeptide, monoclonal antibodies, follistatin, and follistatin-like proteins, have been evaluated for the treatment of muscle-wasting disorders [7, 13, 32–34]. While these studies used recombinant follistatin as myostatin inhibitor, the present study demonstrated that amygdalin in S. Persicae could induce the expression of follistatin mRNA.
Two recent studies applied S. Persicae or S. Persicae-containing formulations (e.g., Fu Zheng Hua Yu recipe) to the treatment of liver fibrosis [35, 36]. Amygdalin in S. Persicae might be helpful in the treatment of fibrosis, according to animal experiments [37]. However, follistatin has recently emerged as an important therapeutic target in the treatment of fibrotic diseases for its potent ability to bind and neutralize activin [14, 17, 38].
Conclusions
Amygdalin isolated from S. Persicae extract in ISF-1 through a bioactivity-guided fractionation procedure induced follistatin expression in HepG2 and C2C12 cell lines.
Abbreviations
- BioReF:
-
Biological response fingerprinting
- EA:
-
Ethyl acetate
- ESI-MS:
-
Electrospray ionization-mass spectrometry
- EtBr:
-
Ethidium bromide
- FDA:
-
U.S. Food and Drug Administration
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- HPLC:
-
High performance liquid chromatography
- ISF-1:
-
Ischemic stroke formulation-1
- LC-MS:
-
Liquid chromatography–mass spectrometry
- mRNA:
-
Messenger RNA
- RP-HPLC:
-
Reverse phase high performance liquid chromatography
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- TGF-β:
-
Transforming growth factor beta
- UV:
-
Ultraviolet.
References
The National Institute of Neurological Disorders and Stroke rt-PA Stroke Study Group: Tissue plasminogen activator for acute ischemic stroke. N Engl J Med. 1995, 333: 1581-1587.
Wardlaw JM, Warlow CP, Counsell C: Systematic review of evidence on thrombolytic therapy for acute ischaemic stroke. Lancet. 1997, 350: 607-614. 10.1016/S0140-6736(97)03022-5.
Scherbakov N, von Haehling S, Anker SD, Dirnagl U, Doehner W: Stroke induced Sarcopenia: muscle wasting and disability after stroke. Int J Cardiol. 2013, 170: 89-94. 10.1016/j.ijcard.2013.10.031.
Dirnagl U: Pathobiology of injury after stroke: the neurovascular unit and beyond. Ann N Y Acad Sci. 2012, 1268: 21-25. 10.1111/j.1749-6632.2012.06691.x.
White TA, Lebrasseur NK: Myostatin and sarcopenia: opportunities and challenges - A mini-review. Gerontology. 2014, 60 (4): 289-93. 10.1159/000356740.
Siriett V, Salerno MS, Berry C, Nicholas G, Bower R, Kambadur R, Sharma M: Antagonism of myostatin enhances muscle regeneration during sarcopenia. Mol Ther. 2007, 15: 1463-1470. 10.1038/sj.mt.6300182.
Whittemore L-A, Song K, Li X, Aghajanian J, Davies M, Girgenrath S, Hill JJ, Jalenak M, Kelley P, Knight A: Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem Biophys Res Commun. 2003, 300: 965-971. 10.1016/S0006-291X(02)02953-4.
Sakuma K, Yamaguchi A: Molecular mechanisms in aging and current strategies to counteract sarcopenia. Curr Aging Sci. 2010, 3: 90-101. 10.2174/1874609811003020090.
Xu L, Zhao P, Mariano A, Han R: Targeted myostatin gene editing in multiple mammalian species directed by a single pair of TALE nucleases. Mol Ther Nucleic Acids. 2013, 2: e112-10.1038/mtna.2013.39.
S-y Y: Inhibins, activins, and follistatins: gonadal proteins modulating the secretion of follicle-stimulating hormone. Endocr Rev. 1988, 9: 267-293. 10.1210/edrv-9-2-267.
Xia Y, Schneyer AL: The biology of activin: recent advances in structure, regulation and function. J Endocrino. 2009, 202: 1-12. 10.1677/JOE-08-0549.
Boström PA, Fernández-Real JM: Metabolism: Irisin, the metabolic syndrome and follistatin in humans. Nat Rev Endocrino. 2014, 10: 11-12.
Rodino Klapac LR, Haidet AM, Kota J, Handy C, Kaspar BK, Mendell JR: Inhibition of myostatin with emphasis on follistatin as a therapy for muscle disease. Muscle Nerve. 2009, 39: 283-296. 10.1002/mus.21244.
Hedger M, de Kretser D: The activins and their binding protein, follistatin—diagnostic and therapeutic targets in inflammatory disease and fibrosis. Cytokine Growth Factor Rev. 2013, 24: 285-295. 10.1016/j.cytogfr.2013.03.003.
Cheng J-C, Chang H-M, Qiu X, Fang L, Leung PC: FOXL2-induced follistatin attenuates activin A-stimulated cell proliferation in human granulosa cell tumors. Biochem Biophys Res Commun. 2014, 44: 537-542.
Rajput SK, Lee K, Zhenhua G, Di L, Folger JK, Smith GW: Embryotropic actions of follistatin: paracrine and autocrine mediators of oocyte competence and embryo developmental progression. Reprod Fertil Dev. 2014, 26: 37-47. 10.1071/RD13282.
Li ZB, Kollias HD, Wagner KR: Myostatin directly regulates skeletal muscle fibrosis. J Biol Chem. 2008, 283: 19371-19378. 10.1074/jbc.M802585200.
Zhu J, Li Y, Lu A, Gharaibeh B, Ma J, Kobayashi T, Quintero AJ, Huard J: Follistatin improves skeletal muscle healing after injury and disease through an interaction with muscle regeneration, angiogenesis, and fibrosis. Am J Pathol. 2011, 179: 915-930. 10.1016/j.ajpath.2011.04.008.
Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, McPherron AC, Wolfman NM, Lee S-J: Induction of cachexia in mice by systemically administered myostatin. Science. 2002, 296: 1486-1488. 10.1126/science.1069525.
Rong J, Tilton R, Shen J, Ng K-M, Liu C, Tam PK-H, Lau AS-Y, Cheng Y-C: Genome-wide biological response fingerprinting (BioReF) of the Chinese botanical formulation ISF-1 enables the selection of multiple marker genes as a potential metric for quality control. J Ethnopharmacol. 2007, 113: 35-44. 10.1016/j.jep.2007.01.021.
C-z H, Wu F, Shen J, Lu L, Fu DL, Liao WJ, Zheng GQ: Clinical efficacy and safety of Buyang Huanwu Decoction for acute ischemic stroke: a systematic review and meta-analysis of 19 randomized controlled trials. Evid Based Complement Alternat Med. 2012, 2012: 630124-
Qi H, Siu SO, Chen Y, Han Y, Chu IK, Tong Y, Lau AS, Rong J: Senkyunolides reduce hydrogen peroxide-induced oxidative damage in human liver HepG2 cells via induction of heme oxygenase-1. Chem Biol Interact. 2010, 183: 380-389. 10.1016/j.cbi.2009.11.029.
Wei L, Liu J, Le XC, Han Y, Tong Y, Lau AS, Rong J: Pharmacological induction of leukotriene B4-12-hydroxydehydrogenase suppresses the oncogenic transformation of human hepatoma HepG2 cells. Int J Oncol. 2011, 39: 735-
Yang CBPW, Zhao J, Cheng YY, Zheng XH, Rong JH: Bornyl caffeate induces apoptosis in human breast cancer MCF-7 cells via the ROS- and JNK-mediated pathways. Acta Pharmacol Sin. 2014, 35: 113-123. 10.1038/aps.2013.162.
Yan J, Tong S, Li J, Lou J: Preparative isolation and purification of amygdalin from prunus armeniaca L. with high recovery by high‒speed countercurrent chromatography. J Liq Chromatogr Relat Technol. 2006, 29 (9): 1271-1279. 10.1080/10826070600598985.
Fukuda T, Ito H, Mukainaka T, Tokuda H, Nishino H, Yoshida T: Anti-tumor promoting effect of glycosides from Prunus persica seeds. Biol Pharm Bull. 2003, 26 (2): 271-273. 10.1248/bpb.26.271.
Kim GJ, Choi HG, Kim JH, Kim SH, Kim JA, Lee SH: Anti-allergic inflammatory effects of cyanogenic and phenolic glycosides from the seed of Prunus persica. Nat Prod Commun. 2013, 8 (12): 1739-1740.
Koo JY, Hwang EY, Cho S, Lee JH, Lee YM, Hong SP: Quantitative determination of amygdalin epimers from armeniacae semen by liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2005, 814 (1): 69-73. 10.1016/j.jchromb.2004.10.019.
Yu L, Ye H, Zheng L, Chen L, Chu K, Liu X, Xu X, Chen G: Determination of the epimerization rate constant of amygdalin by microemulsion electrokinetic chromatography. Electrophoresis. 2011, 32 (2): 218-222. 10.1002/elps.201000287. 38
Kosuge T, Ishida H, Ishii M: Studies on active substances in the herbs used for oketsu (" stagnant blood") in Chinese medicine. II. On the anticoagulative principle in persicae semen. Chem Pharm Bull (Tokyo). 1985, 33: 1496-1498. 10.1248/cpb.33.1496.
Tanaka R, Nitta A, Nagatsu A: Application of a quantitative 1H-NMR method for the determination of amygdalin in Persicae semen, Armeniacae semen, and Mume fructus. J Nat Med. 2014, 68: 225-230. 10.1007/s11418-013-0783-y.
Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore L-A, Ahima RS, Khurana TS: Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002, 420: 418-421. 10.1038/nature01154.
Kim Y, Bobbili N, Paek K, Jin H: Production of a monoclonal anti-myostatin antibody and the effects of in ovo administration of the antibody on posthatch broiler growth and muscle mass. Poult Sci. 2006, 85: 1062-1071. 10.1093/ps/85.6.1062.
Bogdanovich S, Perkins KJ, Krag TO, Whittemore L-A, Khurana TS: Myostatin propeptide-mediated amelioration of dystrophic pathophysiology. FASEB J. 2005, 19: 543-549. 10.1096/fj.04-2796com.
Xie H, Tao Y, Lv J, Liu P, Liu C: Proteomic analysis of the effect of fuzheng huayu recipe on fibrotic liver in rats. Evid Based Complement Alternat Med. 2013, 2013: 972863-
Liu C, Hu Y, Xu L, Liu C, Liu P: Effect of Fuzheng Huayu formula and its actions against liver fibrosis. Chin Med. 2009, 4: 12-10.1186/1749-8546-4-12.
Guo J, Wu W, Sheng M, Yang S, Tan J: Amygdalin inhibits renal fibrosis in chronic kidney disease. Mol Med Reports. 2013, 7: 1453-1457.
Sidis Y, Schneyer AL, Sluss PM, Johnson LN, Keutmann HT: Follistatin: essential role for the N-terminal domain in activin binding and neutralization. J Biol Chem. 2001, 276: 17718-17726. 10.1074/jbc.M100736200.
Acknowledgements
This study was supported by General Research Fund (GRF) (HKU 775812 M) from the Research Grants Council of Hong Kong and the Seed Fund for Basic Research Programme, the University of Hong Kong, offered to JR.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JR and XL conceived and designed the study. CY performed the experiments and the statistical analysis. JR and CY wrote the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Yang, C., Li, X. & Rong, J. Amygdalin isolated from Semen Persicae (Tao Ren) extracts induces the expression of follistatin in HepG2 and C2C12 cell lines. Chin Med 9, 23 (2014). https://doi.org/10.1186/1749-8546-9-23
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1749-8546-9-23