Abstract
Background
The majority of beta-defensin family members are exclusively expressed in the epididymis, and some members have been shown to play essential roles in sperm maturation and fertility in rats, mice and humans. Therefore, beta-defensins are hypothesized to be potential targets for contraception and infertility diagnosis and treatment. Clarifying the regulatory mechanisms for the expression of these genes is necessary. Androgen/androgen receptor (AR) signaling plays an important regulatory role in epididymal structure and function. However, very little is known about the androgenic regulation on the production and secretion of the epididymal beta-defensins.
Methods
The expression of beta-defensins was detected by quantitative RT-PCR. The androgen dependence of beta-defensins was determined by bilateral orchiectomy and androgen supplementation. The androgen response elements (AREs) in the promoters of beta-defensins were identified using the MatInspector software. The binding of AR to AREs was assayed by ChIP-PCR/qPCR.
Results
We demonstrated that 23 mouse caput epididymal beta-defensins were differentially regulated by androgen/androgen receptor. Six genes, Defb18, 19, 20, 39, 41, and 42, showed full regulation by androgens. Ten genes, Defb15, 30, 34, 37, 40, 45, 51, 52, 22 and Spag11a, were partially regulated by androgens. Defb15, 18, 19, 20, 30, 34, 37, 39, 41, 42, 22 and Spag11a were associated with androgen receptor binding sites in their promoter or intronic regions, indicating direct regulation of AR. Six genes, Defb1, 12, 13, 29, 35, and spag11b/c, exhibited an androgen-independent expression pattern. One gene, Defb25, was highly dependent on testicular factors rather on androgens.
Conclusions
The present study provides novel insights into the mechanisms of androgen regulation on epididymal beta-defensins, enabling a better understanding of the function of beta-defensins in sperm maturation and fertility.
Similar content being viewed by others
Background
Beta-defensins are small cationic peptides that exhibit broad-spectrum antimicrobial properties and contribute to mucosal immune responses at epithelial sites. Recently, the complete genome sequences of different species and computational prediction and experimental verification have identified 30–50 novel beta-defensin genes in humans, rats and mice that are organized into gene clusters localized at specific chromosomes [1, 2]. The mRNAs encoding the majority of beta-defensins are exclusively expressed in the epididymis [3, 4]. Epididymal beta-defensins with antimicrobial activity [5–7] have been shown to be involved in epididymal innate immune protection; however, recent studies have indicated that several of these peptides could bind to the sperm surface and play novel roles in male reproductive physiology. Rat Bin1b, which binds to the sperm head, initiates sperm motility in immature sperm from the caput epididymidis by a mechanism dependent on calcium uptake [8]. Likewise, immunization with the Bin1b peptide induced the production of anti-Bin1b antibodies and resulted in reduced fertility in rats [9]. Defb15, which binds to the sperm acrosomal region and forms part of the sperm glycocalyx, is required for sperm motility and male fertility. The in vivo knockdown of the rat Defb15 gene by RNAi led to a considerable attenuation of sperm motility and fertility [7]. Studies that include target deletion of Defb15 have also demonstrated homozygous males with low motility sperm and a reduced fertility phenotype [10]. Recent studies have further enhanced our understanding of the role of beta-defensins in fertility and sterility. Zhou et al. reported that the homozygous deletion of a cluster of nine β-defensin genes (DefbΔ9) in mice resulted in male sterility [11]. Tollner et al. reported that a common mutation in the human defensin Defb126 causes reduced sperm penetration ability and is associated with subfertility [12].
Because of their important functions in sperm maturation and fertility, beta-defensins are receiving more attention and are hypothesized to be potential targets for diagnosing and treating infertility. Therefore, clarifying the regulation mechanisms on their expression is greatly required. However, up to now, very little is known about the regulation of the production and secretion of epididymal beta-defensins.
Androgen signaling plays an important regulatory role in epididymal structure and function. Many epididymal secretory proteins involved in sperm maturation have been identified as androgen-regulated proteins. The effects of androgen are mediated through the androgen receptor (AR), a ligand-inducible nuclear receptor that regulates the expression of target genes by binding to androgen response element (ARE) DNA [13]. The identification of the beta-defensin transcripts that are regulated by androgens might be important to help elucidate the process of sperm maturation. Several beta-defensins have been reported to be regulated by androgen in different species, including rats, monkeys, humans and mice. In mice, only five beta-defensins, including Spag11a (Bin1b), Defb20, 22, 41, and 42 [14–17], have been shown to be regulated by androgen. However, the previous study was unable to determine whether these beta-defensins are direct or indirect AR targets. Additionally, these previous studies were focused on the individual beta-defensin genes; a systematic profile of the androgenic regulation of the beta-defensin family is lacking.
In the epididymis, the majority of beta-defensins are abundantly expressed in the caput region, which expresses high levels of AR and proteins required for sperm maturation. In this study, we systematically investigated androgen regulation on the expression of beta-defensins in the mouse caput epididymidis. Additionally, we screened and identified the AR binding sites associated with some androgen-regulated beta-defensins, indicating direct AR regulation of these defensins.
Methods
Castration and androgen replacement
Adult 10-wk-old male C57BL/6 J mice were divided into the following three groups (six mice/group): an intact group (no castration), an Oil group (castration + oil) and a testosterone propionate (TP) group (castration + TP). The mice were castrated bilaterally under sodium pentobarbital anesthesia. After 7 days of recovery, all of the castrated mice were divided into two groups: the Oil group and the TP group. The Oil group was treated with a 100-μl injection of 90% sesame oil and 10% ethanol. The TP group was injected with 2.5 mg of TP dissolved in 90% sesame oil and 10% ethanol once every 24 h, for a total treatment of 5 mg TP over a 48-h period. The oil and TP were injected into the peritoneal cavity of the mice. All of the mice were killed 48 h post-injection, and the caput epididymides (not including the initial segment) from each group were removed for chromatin immunoprecipitation (ChIP) or real-time polymerase chain reaction (RT-PCR). All of the mouse treatments were repeated three times. The testosterone content of the serum samples was measured using a radioimmunoassay (RIA). The animal experiments were conducted by following a protocol approved by the Institute Animal Care Committee. The protocol conforms to internationally-accepted guidelines for the humane care and use of laboratory animals.
ChIP
Three adult male C57BL/6 J mice from each group were sacrificed, and the caput epididymides were removed, pooled and finely minced in phosphate-buffered saline (PBS). After washing twice with PBS, the tissue debris was cross-linked in 1% formaldehyde at room temperature for 10 min. The debris was pelleted, washed and resuspended in PBS containing protease inhibitors. The debris was then disaggregated (maintaining the samples on ice) using a glass homogenizer with 50 strokes. The nuclei were collected and sonicated to yield 100-500-bp DNA fragments. The lysates were precleared with protein A beads (sc-2001, Santa Cruz, USA) at 4°C for 2 h before an overnight incubation with 4 μg of anti-AR antibody (H-280, sc-13062, Santa Cruz, USA) or normal rabbit IgG (sc-2027, Santa Cruz, USA). After 2 h of incubation with the protein A beads, the chromatin-antibody-beads complex was washed twice with low salt buffer, twice with high salt buffer, twice with lithium chloride buffer and three times with TE buffer. Then, the chromatin was eluted with elution buffer (1% SDS, 0.1 M NaHCO3) before the reversal of cross-linking with proteinase K (Invitrogen, Carlsbad, CA) at 65°C for 4 h. The DNA was purified by two rounds of phenol-chloroform extraction and ethanol precipitation and was resolved in an optimal volume of ddH2O.
RNA extraction and quantitative RT-PCR
The total RNA was extracted from the mouse caput epididymidis (not including the initial segment) using TRIzol (Invitrogen, Grand Island, USA) according to the manufacturer’s instructions. One microgram of total RNA was used to synthesize the first-strand of cDNA with ReverTra Ace reverse transcriptase (Toyobo, Japan) and oligo dT under the conditions recommended by the manufacturer. Quantitative RT-PCR was performed using SYBR Green master mix (Toyobo, Japan) with a Rotor-Gene 3000 machine (Corbett Research, Sydney, Australia). The samples and standard curves were performed in triplicate, and the relative standard curve method was used to calculate the relative gene expression. A housekeeping gene, ribosomal protein S2, was included as the endogenous normalization control to adjust for unequal amounts of RNA. The primers for the defensin genes are listed in Additional file 1: Table S1.
ChIP-PCR and ChIP-qPCR
ChIP assays were performed on mouse caput epididymides (n = 3) as described above. Primers were designed to amplify a 90- to 150-bp region surrounding the known binding peaks. DNA samples from the ChIP preparations were analyzed using conventional PCR or quantitative PCR with SYBR Green Master Mix (Toyobo). The primers are listed in Additional file 2: Table S2.
Bioinformatics
Potential ARE search: The sequences of AR binding sites were scanned for potential ARE motifs using the MatInspector software (Genomatix Software GmbH, Germany) with the default settings. MatInspector utilizes MatBase Matrix Library 8.1, which is a comprehensive transcription factor binding site database that is frequently used to locate DNA sequences. Sequence logos for potential AREs were constructed using Weblogo (http://weblogo.berkeley.edu/logo.cgi) with the default settings.
Genomic mapping of ARBSs: The file containing unique ChIP-seq tags in the WIG format was uploaded to the UCSC genome browser on the mouse mm9 assembly (http://genome.ucsc.edu/cgi-bin/hgGateway) using “add custom tracks”. The distribution of ARBSs around AR target beta-defensin genes was displayed at optimal settings in “configure tracks and display”.
Statistical analyses
Three replicates of all data were performed and used to determine statistical significance. All statistical analyses were carried out with SPSS13.0 (Chicago, IL, USA). Data are expressed as the mean ± SD. For determination of significant differences among groups, one-way analysis of variance was used. P < 0.05 was considered statistically significant in all the tests.
Results
Identification of beta-defensin genes expressed in the caput epididymidis
We focused our study on caput epididymidis and compiled 23 beta-defensin genes expressed in the caput region based on the literature [4, 15, 16, 18, 19]. These genes include Defb1, 12, 13, 15, 18, 19, 20, 22, 25, 29, 30, 34, 35, 37, 39, 40, 41, 42, 45, 51, and 52, Spag11a, and Spag11b/c. The NCBI Gene ID, the RefSeq accession number and the known aliases of these genes are listed in Table 1. Some mouse beta-defensin genes have other aliases or designations; for example, Defb41 is also known as Defb16, Defb19 is also known as Defb24, Defb45 is also known as Defb27, Defb42 is also known as Defb44, and Spag11a is also known as Ep2e, Bin1b or Spag11. In our study, only the official names provided by mouse genome informatics (MGI) were used.
Androgen regulation of beta-defensin gene expression in the mouse caput epididymidis
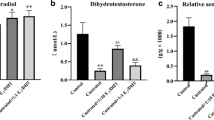
To investigate the transcriptional regulation of beta-defensin genes, we analyzed the expression of these genes in gonadectomized mice. We performed quantitative RT-PCR analyses using total RNAs isolated from the caput epididymides of intact mice and mice that had been bilaterally castrated, reared for 1 week and then treated with TP or oil. As shown in Figure 1A, the expression of 16 genes, including Defb15, 18, 19, 20, 30, 34, 37, 39, 40, 41, 42, 45, 51, 52, and 22 and spag11a, partially or completely disappeared after castration and oil treatment (p < 0.01), whereas after TP treatment, the mRNA level of these genes was significantly increased compared with the mRNA level after oil treatment (p < 0.05), indicating the androgenic regulation of the expression of these genes. The expression levels of 6 genes, Defb18, 19, 20, 39, 41, and 42, were increased and were approximately consistent with the levels observed in the intact mice after TP treatment (p > 0.05), indicating complete regulation by androgens. The expression levels of ten genes, Defb15, 30, 34, 37, 40, 45, 51, 52, and 22 and spag11a, were significantly lower after TP treatment compared with the levels in intact mice, indicating partial regulation by androgens (p < 0.05). Six genes, Defb1, 12, 13, 29, and 35 and spag11b/c, exhibited an androgen-independent expression pattern (Figure 1B). The expression levels of these genes did not change notably with castration and TP treatment, indicating that they are constitutively expressed in the epididymis, regardless of the presence of androgen or testicular factors. One gene, Defb25, was down-regulated by castration, and the expression was not increased after TP treatment, indicating that its expression was highly dependent on testicular factors rather than on androgens (Figure 1B).
The responsiveness of epididymal beta-defensins to androgen manipulation as revealed by quantitative RT-PCR. RNA samples were prepared from the caput epididymides of intact mice (Intact) and mice castrated for 7 days and supplemented with oil (Oil) or testosterone propionate (TP). Different genes showed different responsiveness to androgen. A) The gene expression was fully or partially dependent on androgen. *: P < 0.01 Oil group versus Intact group; **: P < 0.05 TP group versus Oil group. ***: P < 0.05 TP group versus Intact group. B) The expression of Defb25 was dependent on testicular factors and not on androgen, whereas the expression of other genes was independent on testicular factors or androgen. *: P < 0.05 Oil group versus Intact group.
AR direct regulation on target beta-defensins
We previously reported the genome-wide androgen receptor binding sites (ARBSs) in the caput epididymidis using ChIP-seq [14]. Of 16 androgen regulated beta-defensins, 12 had one or more ARBSs within the region 10 kb upstream of the transcription start site to 5 kb downstream of the 3′-end of the gene. As shown in Figure 2 and Table 2, eleven defensin genes were associated with only one ARBS, whereas Defb19 was associated with two ARBSs. One ARBS was simultaneously associated with Defb30 and Defb42, implying the potential dual regulatory functions of this site. The ARBSs associated with Defb18, 19, 20, 30, 34, 41, 42, and 22 and Spag11a were located upstream of the cognate transcription start site, the ARBS associated with Defb15 was located in the downstream of the 3′-end, and the ARBSs associated with Defb37 and Defb39 were located in the intronic region.
Screenshots of the UCSC genome browser mm9 showing AR binding sites around the AR-target beta-defensin genes. The figure was generated by uploading the file containing unique ChIP-seq tags in the WIG format to the UCSC genome browser. The upper blue track shows the tag density, and the AR-target beta-defensin genes are shown at the bottom. The black arrow indicates gene orientation. Sequence positions and other generic UCSC annotations were removed for clarity. One peak formed by a large amount of localized ChIP-seq tags represents an AR binding site. Defb19 was associated with two peaks, whereas Defb30 and Defb42 share one peak. Defb18, 34, 39, 41, 20, 15, 37, and 22 and Spag11a were associated with one peak respectively.
The 12 ARBSs were searched for potential AREs. As shown in Table 2, the 10 ARBSs associated with Defb15, 18, 19, 34, 37, 39, 41, and 22 and Spag11a contained one or more potential AREs, whereas the 2 ARBSs associated with Defb20, 30 and 42 were not potential AREs. The sequence logo for the potential ARE motif overrepresented in the above-mentioned 10 ARBSs was generated based on sequence similarity. As shown in Additional file 3: Figure S1, the identified potential ARE motif from 10 ARBSs (top panel) was very similar to the established consensus canonical ARE in the MatBase database (bottom panel).
To validate the ChIP-seq results, the ARBSs associated with androgen-regulated beta-defensins were confirmed by individual ChIP assays combined with conventional PCR (ChIP-PCR) or quantitative PCR (ChIP-qPCR). As shown in Figure 3A, all of the ARBSs were occupied by AR in the caput epididymidis under physiological conditions. Given that AR recruitment to target DNA is androgen-dependent, we assessed the androgen dependence of AR recruitment to ARBSs using ChIP-qPCR. AR binding was abolished after castration and recovered following androgen supplementation (Figure 3B), indicating that AR occupancy is androgen dependent.
AR occupancies on epididymal AR binding sites associated with androgen-regulated beta-defensins as revealed by ChIP-PCR (A) or ChIP-qPCR (B). A) Input sample (IP) and ChIP samples of anti-AR antibody (AR) or normal IgG (IgG) were analyzed using PCR to confirm AR binding at eleven epididymal target loci. B) ChIP samples were prepared from the caput epididymidis of intact mice (Intact) and mice castrated for 7 days and supplemented with oil (Oil) or testosterone propionate (TP) for an additional 2 days. Enrichment folds were calculated using IgG enrichment as a control. The data are presented as the mean ± SD of three replicates.
Discussion
The epididymis is an androgen-responsive organ responsible for sperm maturation and storage. Compared with other hormones and testicular factors, androgens play important regulatory roles in epididymal structure and function [20]. There is a high level of androgen receptors in the murine epididymis, and because of the importance of the epididymidis in the regulation of sperm maturation, the identification of the transcripts regulated by androgens might facilitate the elucidation of the process of sperm maturation [21].
Many members of the beta-defensin family have shown exclusive expression in the epididymis. Except for the 6-cysteine domain, which is rich in positively charged amino acids, beta-defensins differ considerably in their amino acid sequences and target pathogen specificity. They form an important component of the epididymal innate immune system and provide protection against a broad range of pathogens that could invade the male reproductive tract, including E. coli, S. aureus and C. albicans [5–7]. Recent research has identified additional functions for beta-defensins that link the innate and adaptive immune responses. Beta-defensins cross-talk with the adaptive immune system by interacting with specific chemokine and toll-like receptors on immature dendritic cells and memory T cells resulting in the modulation of immunocompetent cell responses of the host [22].
Epididymal beta-defensins have evolved to perform functions other than immunoprotection, such as promoting sperm maturation, as suggested by the presence of several defensins on the sperm surface. However, little is known about the hormonal regulation, especially androgenic regulation, of the expression of beta-defensins.
In this study, androgen and AR regulation on caput epididymal beta-defensins was investigated. Beta-defensins exhibited different responses to androgen manipulation; most were regulated by androgen, some were not regulated by androgen or testicular factors, and one, Defb25, was regulated by testicular factors and not by androgen.
Among the androgen regulated beta-defensin genes, Defb20, 41, 42 and Spag11a were previously reported to be regulated by androgen [14–16]; our study further verified that these genes were direct targets of AR and had AR binding sites in the proximal promoter regions. Defb30 was previously shown to be regulated by testicular factors and not by androgens [16]. In our study, Defb30 was partially regulated by androgen, and it could be a direct target of AR. This discordance might be attributed to the sensitivity of different methods. The previous study used Northern blot to detect the expression of Defb30, whereas the current study used quantitative RT-PCR. Compared with quantitative RT-PCR, Northern blot has a low sensitivity. We reanalyzed the previous data and found a weak staining signal in the androgen replacement mouse epididymis compared with the castrated mouse epididymis, although the authors ignored it.
Jalkanen et al. reported that Defb42 was partially regulated by androgen [15], whereas Oh et al. reported that Defb42 was regulated by testicular factors and not by androgen [16]. Our results demonstrated that Defb42 was regulated by androgens. To investigate this discrepancy, we reanalyzed the data from Oh et al. and determined that the Defb42 in their paper should be classified as Defb48 based on sequence alignment. Oh et al. also reported that Defb44 was regulated by androgen; however, in that paper, Defb44 should be classified as Defb42 based on the sequence alignment.
AR regulates the expression of target genes by binding to AREs in the genome. Our previous work has identified genome-wide AR binding sites in the mouse epididymis, and in this study, we screened and identified ARBSs associated with androgen-regulated beta-defensins. These sites were located in the proximal promoters or intronic regions of target genes, indicating that their expression is directly regulated by AR. Consistent with our expectations, potential ARE motifs were identified in the majority of ARBSs, and these motifs conformed to consensus sequences that were highly similar to the established ARE in the MatBase database. However, no potential AREs were identified in the two ARBSs associated with Defb20 and Defb30/42, suggesting AR regulation by a tethering mechanism. AR regulation occurs without the direct binding of AR to AREs and likely through tethered associations of the receptor with its target genes [23]. For the ARBS associated with Defb30/42, an AP-2 motif was found in the binding summit. AP-2 has been reported to be an AR coregulator in the mouse caput epididymidis [14, 24]. Thus, AR might be recruited to the ARBS by interacting with AP-2. Another possibility that could not be excluded is that there could be AREs that do not conform to the consensus sequence in the ARBSs associated with Defb20 and Defb30/42. For example, our previous data showed that noncanonical or half-site-like AREs were enriched in 38.3% of ARBSs without canonical AREs in the mouse epididymis. Those genes that changed expression, although were not bound by an AR, might be indirect AR target genes or might be regulated via long-range regulation (binding outside our defined window).
We tested the androgen responsiveness of Defb22, which is the mouse orthologue of human Defb126, and determined that it was partially regulated by androgen. Our ChIP-qPCR data identified an AR binding site in the distal promoter region, suggesting that Defb22 was a direct target of AR. Among the 9 genes deleted by Zhou et al. [11], four (Defb1, 13, 15, and 35) were tested in our study. Defb15 was partially regulated by androgen, whereas the three other defensins (Defb1, 13, 35) were not regulated by androgen.
The present study provides novel insights into the mechanisms of androgen regulation on epididymal beta-defensins. Sperm motility and fertilization capacity have been reported to be regulated by androgen [20]. Some AR-target beta-defensins, such as Spag11a, Defb22 and Defb15, might be involved in this regulatory process. Further studies are needed to clarify the functions of all of the AR target beta-defensins to help elucidate the molecular mechanism of sperm maturation and fertility.
Conclusions
The current study demonstrated that 23 beta-defensins from caput epididymis showed different responsiveness to androgen, and twelve of them were identified to be directly regulated by androgen receptor. These results shed new light on the mechanisms of androgen/AR regulation on epididymal gene expression.
Abbreviations
- AR:
-
Androgen receptor
- ARE:
-
Androgen response element
- ARBS:
-
Androgen receptor binding site
- ChIP:
-
Chromatin immunoprecipitation
- MGI:
-
Mouse genome informatics
- RIA:
-
Radioimmunoassay
- TP:
-
Testosterone propionate.
References
Patil AA, Cai Y, Sang Y, Blecha F, Zhang G: Cross-species analysis of the mammalian beta-defensin gene family: presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiol Genomics. 2005, 23 (1): 5-17.
Schutte BC, Mitros JP, Bartlett JA, Walters JD, Jia HP, Welsh MJ, Casavant TL, McCray PB: Discovery of five conserved beta -defensin gene clusters using a computational search strategy. Proc Natl Acad Sci U S A. 2002, 99 (4): 2129-2133.
Yamaguchi Y, Nagase T, Makita R, Fukuhara S, Tomita T, Tominaga T, Kurihara H, Ouchi Y: Identification of multiple novel epididymis-specific beta-defensin isoforms in humans and mice. J Immunol. 2002, 169 (5): 2516-2523.
Zaballos A, Villares R, Albar JP, Martinez AC, Marquez G: Identification on mouse chromosome 8 of new beta-defensin genes with regionally specific expression in the male reproductive organ. J Biol Chem. 2004, 279 (13): 12421-12426.
Diao H, Guo C, Lin D, Zhang Y: Intein-mediated expression is an effective approach in the study of beta-defensins. Biochem Biophys Res Commun. 2007, 357 (4): 840-846.
Yenugu S, Hamil KG, Radhakrishnan Y, French FS, Hall SH: The androgen-regulated epididymal sperm-binding protein, human beta-defensin 118 (DEFB118) (formerly ESC42), is an antimicrobial beta-defensin. Endocrinology. 2004, 145 (7): 3165-3173.
Zhao Y, Diao H, Ni Z, Hu S, Yu H, Zhang Y: The epididymis-specific antimicrobial peptide beta-defensin 15 is required for sperm motility and male fertility in the rat (Rattus norvegicus). Cell Mol Life Sci. 2010, 68 (4): 697-708.
Zhou CX, Zhang YL, Xiao L, Zheng M, Leung KM, Chan MY, Lo PS, Tsang LL, Wong HY, Ho LS, Chung YW, Chan HC: An epididymis-specific beta-defensin is important for the initiation of sperm maturation. Nat Cell Biol. 2004, 6 (5): 458-464.
Xu W, Zhang X, Chen W, Fok KL, Rowlands DK, Chui YL, Chan HC: Immunization with Bin1b decreases sperm motility with compromised fertility in rats. Fertil Steril. 2010, 93 (3): 952-958. e951
Semple F, Dorin JR: Beta-Defensins: multifunctional modulators of infection, inflammation and more?. J Innate Immun. 2012, 4 (4): 337-348.
Zhou YS, Webb S, Lettice L, Tardif S, Kilanowski F, Tyrrell C, Macpherson H, Semple F, Tennant P, Baker T, Hart A, Devenney P, Perry P, Davey T, Barran P, Barratt CL, Dorin JR: Partial deletion of chromosome 8 beta-defensin cluster confers sperm dysfunction and infertility in male mice. PLoS Genet. 2013, 9 (10): e1003826-
Tollner TL, Venners SA, Hollox EJ, Yudin AI, Liu X, Tang G, Xing H, Kays RJ, Lau T, Overstreet JW, Xu X, Bevins CL, Cherr GN: A common mutation in the defensin DEFB126 causes impaired sperm function and subfertility. Sci Transl Med. 2011, 3 (92): 92ra65-
Patrao MT, Silva EJ, Avellar MC: Androgens and the male reproductive tract: an overview of classical roles and current perspectives. Arq Bras Endocrinol Metabol. 2009, 53 (8): 934-945.
Hu S, Yao G, Guan X, Ni Z, Ma W, Wilson EM, French FS, Liu Q, Zhang Y: Research resource: Genome-wide mapping of in vivo androgen receptor binding sites in mouse epididymis. Mol Endocrinol. 2010, 24 (12): 2392-2405.
Jalkanen J, Huhtaniemi I, Poutanen M: Discovery and characterization of new epididymis-specific beta-defensins in mice. Biochim Biophys Acta. 2005, 1730 (1): 22-30.
Oh J, Lee J, Woo JM, Choi E, Park I, Han C, Baek N, Lee H, Kim do H, Cho C: Systematic identification and integrative analysis of novel genes expressed specifically or predominantly in mouse epididymis. BMC genomics. 2006, 7: 314-
Pujianto DA, Loanda E, Sari P, Midoen YH, Soeharso P: Sperm-associated antigen 11A is expressed exclusively in the principal cells of the mouse caput epididymis in an androgen-dependent manner. Reprod Biol Endocrinol. 2013, 11: 59-
Johnston DS, Jelinsky SA, Bang HJ, DiCandeloro P, Wilson E, Kopf GS, Turner TT: The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod. 2005, 73 (3): 404-413.
Johnston DS, Turner TT, Finger JN, Owtscharuk TL, Kopf GS, Jelinsky SA: Identification of epididymis-specific transcripts in the mouse and rat by transcriptional profiling. Asian J Androl. 2007, 9 (4): 522-527.
Robaire B, Seenundun S, Hamzeh M, Lamour SA: Androgenic regulation of novel genes in the epididymis. Asian J Androl. 2007, 9 (4): 545-553.
Chauvin TR, Griswold MD: Androgen-regulated genes in the murine epididymis. Biol Reprod. 2004, 71 (2): 560-569.
Semple CA, Rolfe M, Dorin JR: Duplication and selection in the evolution of primate beta-defensin genes. Genome Biol. 2003, 4 (5): R31-
Patki M, Chari V, Sivakumaran S, Gonit M, Trumbly R, Ratnam M: The ETS domain transcription factor ELK1 directs a critical component of growth signaling by the androgen receptor in prostate cancer cells. J Biol Chem. 2013, 288 (16): 11047-11065.
Pihlajamaa P, Sahu B, Lyly L, Aittomäki V, Hautaniemi S, Jänne OA: Tissue-specific pioneer factors associate with androgen receptor cistromes and transcription programs. EMBO J. 2014, 33 (4): 312-326.
Acknowledgements
The manuscript was edited for proper English language by American Journal Experts (Certificate Verification Key: 4C15-7185-7F5E-98A6-D1A6). This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81200468, 81370762), the Key program for Basic Research of the Science and Technology Commission of Shanghai Municipality, China (Grant No.12JC1405800), The National Basic Research Program (973 Program) (Grant No. 2014CB943300), Research special fund of national health and family planning commission, China (Grant No. 201402004).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SGH performed quantitative RT-PCR, data analysis and wrote the manuscript. MZ performed literature research and statistical analysis. GXY performed ChIP assays while WBM and XQL constructed the mouse castration model. QLZ performed bioinformatics analysis. ZJC and YS designed the study and took part in the manuscript preparation. All authors read and approved the final manuscript.
Electronic supplementary material
12958_2014_1245_MOESM3_ESM.tiff
Additional file 3: Figure S1: The potential ARE sequence (upper panel) in ARBSs associated with androgen-regulated beta-defensins identified by Weblogo is very similar to the ARE in the MatBase database (bottom panel). (TIFF 1 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Hu, SG., Zou, M., Yao, GX. et al. Androgenic regulation of beta-defensins in the mouse epididymis. Reprod Biol Endocrinol 12, 76 (2014). https://doi.org/10.1186/1477-7827-12-76
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7827-12-76