Abstract
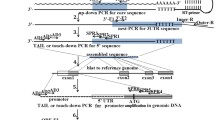
'New RACE' (rapid amplification of cDNA ends) PCR is a method for obtaining full-length cDNA for mRNA for which only part of the sequence is known. Starting with cellular mRNA, PCR is used to amplify regions between the known parts of the sequence and nonspecific tags at the ends of the cDNA. In 'new RACE', an anchor is ligated to the 5′ end of the mRNA before reverse transcription, resulting in the selective production of full-length 5′ cDNA ends. Although 'new RACE' can also be used to amplify 3′ ends, only the protocol for obtaining 5′ ends is presented here. This protocol can be completed in 1–3 days.

Similar content being viewed by others
References
Liu, X. & Gorovsky, M.A. Mapping the 5′ and 3′ ends of tetrahymena-thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res. 21, 4954–4960 (1993).
Fromont-Racine, M., Bertrand, E., Pictet, R. & Grange, T. A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res. 21, 1683–1684 (1993).
Scotto-Lavino, E., Du, G. & Frohman, M.A. 5′ end cDNA amplification using Classic RACE. Nat. Protocols 1, 2555–2562 (2006).
Volloch, V., Schweitzer, B., Zhang, X. & Rits, S. Identification of negative-strand complements to cytochrome oxidase subunit III RNA in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 88, 10671–10675 (1991).
Mandl, C.W., Heinz, F.X., Puchhammer-Stockl, E. & Kunz, C. Sequencing the termini of capped viral RNA by 5′-3′ ligation and PCR. Biotechniques 10, 484–486 (1991).
Tessier, D.C., Brousseau, R. & Vernet, T. Ligation of single-stranded oligodeoxyribonucleotides by T4 RNA ligase. Anal. Biochem. 158, 171–178 (1986).
Frohman, M.A., Dickinson, M.E., Hogan, B.L.M. & Martin, G.R. Localization of two new and related homeobox-containing genes to chromosomes 1 and 5, near the phenotypically similar mutant loci dominant hemimelia (Dh) and hemimelic extra-toes (Hx). Mouse Genome 91, 323–325 (1993).
Don, R.H., Cox, P.T., Wainwright, B.J., Baker, K. & Mattick, J.S. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19, 4008 (1991).
Frohman, M.A. On beyond classic RACE (rapid amplification of cDNA ends). PCR Methods Appl. 4, S40–S58 (1994).
Scotto-Lavino, E., Du, G. & Frohman, M.A. 3′ end cDNA amplification using classic RACE. Nat. Protocols 1, 2742–2745 (2006).
Zhang, Y. in Generation of cDNA Libraries (ed. Ying, S.-Y.) 13–24 (Humana Press, Totowa, New Jersey, 2003).
Acknowledgements
We thank Y. Zhang and Humana Press for permission to use and adapt material from a prior review (ref. 11). Supported by the National Institutes of Health (NIHDDK 64166 and NIHGM71520 to M.A.F., and NIHGM071475 to G.D.) and the American Heart Association (Scientist Development Grant (0430096N to G.D.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Scotto-Lavino, E., Du, G. & Frohman, M. Amplification of 5′ end cDNA with 'new RACE'. Nat Protoc 1, 3056–3061 (2006). https://doi.org/10.1038/nprot.2006.479
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.479
- Springer Nature Limited
This article is cited by
-
A versatile 5′ RACE-Seq methodology for the accurate identification of the 5′ termini of mRNAs
BMC Genomics (2022)
-
Genome-wide quantification of 5′-phosphorylated mRNA degradation intermediates for analysis of ribosome dynamics
Nature Protocols (2016)
-
Focused transcription from the human CR2/CD21 core promoter is regulated by synergistic activity of TATA and Initiator elements in mature B cells
Cellular & Molecular Immunology (2016)
-
An efficient full-length cDNA amplification strategy based on bioinformatics technology and multiplexed PCR methods
Scientific Reports (2016)
-
Diversity of transcripts and transcript processing forms in plastids of the dinoflagellate alga Karenia mikimotoi
Plant Molecular Biology (2016)