Abstract
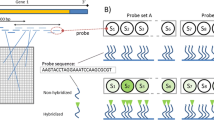
Molecular barcode arrays allow the analysis of thousands of biological samples in parallel through the use of unique 20-base-pair (bp) DNA tags. Here we present a new barcode array, which is unique among microarrays in that it includes at least five replicates of every tag feature. The use of smaller dispersed replicate features dramatically improves performance versus a single larger feature and allows the correction of previously undetectable hybridization defects.



Similar content being viewed by others
References
Winzeler, E.A. et al. Science 285, 901–906 (1999).
Giaever, G. et al. Nature 418, 387–391 (2002).
Lum, P.Y. et al. Cell 116, 121–137 (2004).
Ooi, S.L., Shoemaker, D.D. & Boeke, J.D. Science 294, 2552–2556 (2001).
Birrell, G.W., Giaever, G., Chu, A.M., Davis, R.W. & Brown, J.M. Proc. Natl. Acad. Sci. USA 98, 12608–12613 (2001).
Birrell, G.W. et al. Proc. Natl. Acad. Sci. USA 99, 8778–8783 (2002).
Steinmetz, L.M. et al. Nat. Genet. 31, 400–404 (2002).
Giaever, G. Trends Pharmacol. Sci. 24, 444–446 (2003).
Lee, W. et al. PLoS Genet. 1, e24 (2005).
Hardenbol, P. et al. Nat. Biotechnol. 21, 673–678 (2003).
Fischer, K.D. et al. Nature 374, 474–477 (1995).
Paddison, P.J. et al. Nature 428, 427–431 (2004).
Shoemaker, D.D., Lashkari, D.A., Morris, D., Mittmann, M. & Davis, R.W. Nat. Genet. 14, 450–456 (1996).
Giaever, G. et al. Nat. Genet. 21, 278–283 (1999).
Eason, R.G. et al. Proc. Natl. Acad. Sci. USA 101, 11046–11051 (2004).
Acknowledgements
This work was supported by a grant from the US National Human Genome Research Institute. We thank B. St. Onge, M. Hillenmeyer and S. Brachat for suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Use of unassigned tag probes to estimate background hybridization. (PDF 136 kb)
Supplementary Figure 2
Effect of repairs on tag performance. (PDF 237 kb)
Supplementary Table 1
Detailed information on the repaired tags. (PDF 160 kb)
Rights and permissions
About this article
Cite this article
Pierce, S., Fung, E., Jaramillo, D. et al. A unique and universal molecular barcode array. Nat Methods 3, 601–603 (2006). https://doi.org/10.1038/nmeth905
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth905
- Springer Nature America, Inc.
This article is cited by
-
Select microtubule inhibitors increase lysosome acidity and promote lysosomal disruption in acute myeloid leukemia (AML) cells
Apoptosis (2015)
-
Dynamic characterization of growth and gene expression using high-throughput automated flow cytometry
Nature Methods (2014)
-
Mitochondrial protein sorting as a therapeutic target for ATP synthase disorders
Nature Communications (2014)
-
Dissecting a complex chemical stress: chemogenomic profiling of plant hydrolysates
Molecular Systems Biology (2013)
-
Target identification and mechanism of action in chemical biology and drug discovery
Nature Chemical Biology (2013)