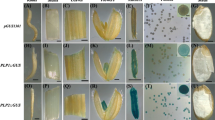
Abstract
The tomato lat52 gene encodes an essential cysteine-rich protein preferentially transcribed in the vegetative cell during pollen maturation. Detailed analyses of the identity, organization and role of cis-regulatory elements in controlling the precise developmental and tissue-specific expression of lat52 during pollen development were performed. Analysis of a series of 5′ promoter deletion mutants stably introduced into tobacco demonstrated differential developmental activation of deletion mutants during pollen development. All major cis-regulatory elements required for pollen-specific transcription were located within the upstream region −492 to −52. This region was shown to comprise three independent activator domains A, B and C, each sufficient to activate the minimal CaMV 35S promoter in a pollen-specific manner. 5′ deletion and gain of function approaches were used to show that domain A and the previously defined motif PBII (sub-domain B1) were largely redundant in the presence of downstream sequences in mature pollen. Within domain B two novel pollen-specific sub-domains B2 and B3 were identified. Within domain C, the activity of the PBI motif (sub-domain C1) was shown to be strictly dependent upon a downstream 20 bp pollen-specific activator unit −72 to −52 (sub-domain C2), containing two novel co-dependent regulatory elements AGAAA and TCCACCATA. These results demonstrate that transcriptional activation of lat52 is controlled by a complex of pollen-specific cis-regulatory elements which cooperate to achieve maximum levels of gene expression throughout pollen maturation. Alternative models of the interaction of identified cis-regulatory elements with putative trans-acting factors within the lat52 promoter and their developmental utilization are presented.
Similar content being viewed by others
References
Batanero E, Villalba M, Lopez-Otin C, Rodriguez R: Isolation and characterization of an olive allergenlike protein from lilac pollen. Sequence analysis of three cDNA encoding protein isoforms. Eur J Biochem 221: 187–193 (1994).
Bate N, Spurr C, Foster G, Twell D: Maturationspecific translational enhancement mediated by the 50UTR of a late pollen transcript. Plant J 10: 613–623 (1996).
Bevan M: Binary Agrobacterium vectors for plant transformation. Nucl Acids Res 12: 8711–8721 (1984).
Buchel AS, Molenkamp R, Bol JF, Linthorst HJM: The PR1a promoter contains a number of elements that bind GT1 like nuclear factors with different affinity. Plant Mol Biol 30: 493–504 (1996).
Carrington JC, Freed DD: Cap-independent enhancement of translation by a plant potyvirus 50 nontranslated region. J Virol 64: 1590–1597 (1990).
Eady C, Lindsey K, Twell D: Differential activation and conserved vegetative cellspecific activity of a late pollen promoter in species with biand tricellular pollen. Plant J 5: 543–550 (1994).
Eyal Y, Curie C, McCormick S: Pollen specificity elements reside in 30 bp of the proximal promoters of two pollenexpressed genes. Plant Cell 7: 373–384 (1995).
Gilmartin PM, Memelink J, Hiratsuka K, Kay SA, Chua NH: Characterization of a gene encoding a DNA binding protein with specificity for a lightresponsive element. Plant Cell 4: 839–849 (1992).
Green PJ, Yong MH, Cuozzo M, Kano-Murakami Y, Silverstein P, Chua NH: Binding site requirements for pea nuclear protein factor GT1 correlate with sequences required for lightdependent transcriptional activation of the rbcS3A gene. EMBO J 7: 4035–4044 (1988).
Guerrero FD, Crossland L, Smutzer GS, Hamilton DA, Mascarenhas JP: Promoter sequences from a maize pollenspecific gene direct transcription in tobacco. Mol Gen Genet 224: 161–168 (1990).
Hamilton DA, Roy M, Rueda J, Sindhu RK, Sanford J, Mascarenhas JP: Dissection of a pollenspecific promoter from maize by transient transformation assays. Plant Mol Biol 18: 211–218 (1992).
Hanson DD, Hamilton DA, Travis JL, Bashe DM, Mascarenhas JP: Characterization of a pollenspecific cDNA clone from Zea mays and its expression. Plant Cell 1: 173–179 (1989).
Hochstenbach R, de Groot P, Jacobs J, Schrauwen J, Wullens G: The promoter of a gene that is expressed only in pollen interacts with ubiquitous transcription factors. Sex Plant Reprod 9: 197–202 (1996).
Jacobsen K, Laursen NB, Jensen EO, Marcker A, Poulsen C, Marcker KA: HMG Ilike proteins from leaf and nodule nuclei interact with different ATmotifs in soybean nodulin promoters. Plant Cell 2: 85–94 (1990).
Jefferson RA, Kavanagh TA, Bevan MW: GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 (1987).
Kuhlemeier C, Cuozzo M, Green PJ, Goyvaerts E, Ward K, Chua NH: Localisation and conditional redundancy of regulatory elements in rbcS3A, a pea gene encoding the small subunit of ribulosebisphosphate carboxylase. Proc Natl Acad Sci USA 85: 4662–4666 (1988).
Lombardero M, Barbas JA, Moscoso del Prado J, Carreira J: cDNA sequence analysis of the main olive allergen, Ole e I. Clin Exp Allergy 24: 765–770 (1994).
McCormick S, Smith A, Gasser C, Sachs K, Hinchee M, Horsch R, Fraley R: The identification of genes specifically expressed in reproductive organs of tomato. In: Nevins D, Jones R (eds) Tomato Biotechnology, pp. 255–265. Alan R. Liss, New York (1987).
McCormick S, Twell D, Vancanneyt G, Yamaguchi J: Molecular analysis of gene regulation and function duringmale gametophyte development. In: Jenkins G, Schuch W (eds) Molecular Biology of Plant Development. Proceedings of SEB symposium No. 45. Company of Biologists, Cambridge (1991).
Muschietti J, Dircks L, Vancanneyt G, McCormick S: LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J 6: 321–338 (1994).
Ow DW, Wood KV, DeLuca M, DeWet JR, Helinski DR, Howell SH: Transient and stable expression of the firefly luciferase gene in plant cells and trangenic plants. Science 234: 856–859 (1986).
Pe ME, Frova C, Colombo L, Binelli G, Mastroianni N, Tarchini R, Visioli G: Molecular cloning of genes expressed in pollen of Sorghum bicolor. Maydica 39: 107–113 (1994).
Perisic O, Lam E: A tobacco DNA binding protein that interacts with a lightresponsive box II element. Plant Cell 4: 831–838 (1992).
Rodriguez-Garcia MI, Fernandez MC, Alche JD: Immunocytochemical localisation of allergenic protein (Ole e I) in the endoplasmic reticulum of the developing pollen grain of olive (Oleo europaea L.). Planta 196: 558–563 (1995).
Schrauwen JAM, de Groot PFM, van Herpen MMA, van der Lee T, Reynen WH, Weterings KAP, Wullems GJ: Stage-related expression of mRNAs during pollen development in lily and tobacco. Planta 182: 298–304 (1990).
Tebbutt SJ, Lonsdale DM: Deletion analysis of a tobacco pollenspecific polygalacturonase promoter. Sex Plant Reprod 8: 242–246 (1995).
Topfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH: A set of plant expression vectors for transcriptional and translational fusions. Nucl Acids Res 15: 5890 (1987).
Tupy J, Ribhova L, Zarsky V: Production of fertile tobacco pollen from microspores in suspension culture and its storage for in situ pollination. Sex Plant Reprod 4: 284–287 (1991).
Turner R, Bate N, Twell D, Foster G: In vivo characterisation of a translational enhancer upstream from the coat protein open reading frame of potato virus S. Arch Virol 137: 123–132 (1994).
Twell D: The diversity and regulation of gene expression in the pathway of male gametophyte development. In: Scott RJ, Stead AD (eds) Molecular and CellularAspects of Plant Reproduction, pp. 83–115. Cambridge University Press, Cambridge (1994).
Twell D: Diptheria toxinmediated cell ablation in developing pollen: vegetative cell ablation blocks generative cell migration. Protoplasma 187: 144–154 (1995).
Twell D, Patel S, Sorenson A, Roberts M, Scott R, Draper J, Foster G: Activation and developmental regulation of an Arabidopsis antherspecific promoter in microspores and pollen of Nicotiana tabacum. Sex Plant Reprod 6: 217–224 (1993).
Twell D, Wing R, Yamaguchi J, McCormick S: Isolation and expression of an antherspecific gene from tomato. Mol Gen Genet 217: 240–245 (1989).
Twell D, Klein TM, Fromm ME, McCormick S: Transient expression of chimeric genes delivered into pollen by microprojectile bombardment. Plant Physiol 91: 1270–1274 (1989).
Twell D, Yamaguchi J, McCormick S: Pollenspecific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109: 705–713 (1990).
Twell D, Yamaguchi J, Wing RA, Ushiba J, McCormick S: Promoter analysis of three genes that are coordinately expressed during pollen development reveals pollenspecific enhancer sequences and shared regulatory elements. Genes Devel 5: 496–507 (1991).
Ursin VM, Yamaguchi J, McCormick S: Gametophytic and sporophytic expression of antherspecific genes in developing tomato anthers. Plant Cell 1: 727–736 (1989).
Villalba M, Batanero E, Lopez-Otin C, Sanchez LM, Monsalve RI, Gonzalez de la Pena MA, Lahoz C, Rodriguez R: The amino acid sequence of Ole e I, the major allergen from olive tree (Olea europaea) pollen. Eur J Biochem 216: 863–869 (1993).
Willing RP, Bashe D, Mascharenhas JP: An analysis of the quantity and diversity of messenger RNAs from pollen and shoots of Zea mays. Theor Appl Genet 75: 751–753 (1988).
Willing RP, Mascharenhas JP: Analysis of the complexity and diversity of mRNAs from pollen and shoots of Tradescantia. Plant Physiol 75: 865–868 (1984).
Zappavigna V, Falciola L, Citterich MH, Mavilio F, Bianchi ME: HMG1 interacts with HOX proteins and enhances their binding and transcriptional activation. EMBO J 15: 4981–4991 (1996).
Zou JT, Zhan XY, Wu HM, Wang H, Cheung AY: Characterisation of a rice pollenspecific gene and its expression. American Journal of Botany 81: 552–561 (1994).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bate, N., Twell, D. Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol 37, 859–869 (1998). https://doi.org/10.1023/A:1006095023050
Issue Date:
DOI: https://doi.org/10.1023/A:1006095023050