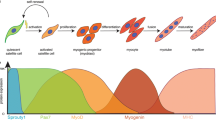
Abstract
Limb regeneration is the outcome of a complex sequence of events that are mediated by interactions between cells derived from the tissues of the amputated stump. Early in regeneration, these interactions are mediated by growth factor/morphogen signaling associated with nerves and the wound epithelium. One shared property of these proregenerative signaling molecules is that their activity is dependent on interactions with sulfated glycosaminoglycans (GAGs), heparan sulfate proteoglycan (HSPG) in particular, in the extracellular matrix (ECM). We hypothesized that there are cells in the axolotl that synthesize specific HSPGs that control growth factor signaling in time and space. In this study we have identified a subpopulation of cells within the ECM of axolotl skin that express high levels of sulfated GAGs on their cell surface. These cells are dispersed in a grid-like pattern throughout the dermis as well as the loose connective tissues that surround the tissues of the limb. These cells alter their morphology during regeneration, and are candidates for being a subpopulation of connective tissue cells that function as the cells required for pattern-formation during regeneration. Given their high level of HSPG expression, their stellate morphology, and their distribution throughout the loose connective tissues, we refer to these as the positional information GRID (Groups that are Regenerative, Interspersed and Dendritic) cells. In addition, we have identified cells that stain for high levels of expression of sulfated GAGs in mouse limb connective tissue that could have an equivalent function to GRID cells in the axolotl. The identification of GRID cells may have important implications for work in the area of Regenerative Engineering.





Similar content being viewed by others
References
Makanae A, Mitogawa K, Satoh A. Co-operative bmp- and Fgf-signaling inputs convert skin wound healing to limb formation in urodele amphibians. Dev Biol. 2014;396:57–66.
McCusker C, Bryant SV, Gardiner DM. The axolotl limb blastema: cellular and molecular mechanisms driving blastema formation and limb regeneration in tetrapods. Regeneration (Oxf). 2015;2:54–71.
Vieira WA, Wells KM, Raymond MJ, De Souza L, Garcia E, McCusker CD. FGF, BMP, and RA signaling are sufficient for the induction of complete limb regeneration from non-regenerating wounds on Ambystoma mexicanum limbs. Dev Biol. 2019;451:146–57.
Bryant SV, Gardiner DM. The relationship between growth and pattern formation. Regeneration (Oxf). 2016;3:103–22.
Nacu E, Gromberg E, Oliveira CR, Drechsel D, Tanaka EM. FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature. 2016;2016(533):407–10.
Phan AQ, Lee J, Oei M, Flath C, Hwe C, Mariano R, et al. Positional information in axolotl and mouse limb extracellular matrix is mediated via heparan sulfate and fibroblast growth factor during limb regeneration in the axolotl (Ambystoma mexicanum). Regeneration (Oxf). 2015;2:182–201.
McCusker CD, Gardiner DM. Positional information is reprogrammed in blastema cells of the regenerating limb of the axolotl (Ambystoma mexicanum). Tsonis PA, editor. PLoS ONE. Public Library of Science; 2013;8:e77064.
McCusker CD, Gardiner DM. Understanding positional cues in salamander limb regeneration: implications for optimizing cell-based regenerative therapies. Dis Model Mech Company of Biologists. 2014;7:593–9.
Nacu E, Glausch M, Le HQ, Damanik FF, Schuez M, Knapp D, et al. Connective tissue cells, but not muscle cells, are involved in establishing the Proximo-distal outcome of limb regeneration in the axolotl. Development. 2013;140:513–8.
Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, et al. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. 2018;362:eaaq0681.
Leigh ND, Dunlap GS, Johnson K, Mariano R, Oshiro R, Wong AY, et al. Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat Commun. 2018;9:5153.
Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–5.
McCusker CD, Diaz-Castillo C, Sosnik J, Q Phan A, Gardiner DM. Cartilage and bone cells do not participate in skeletal regeneration in Ambystoma mexicanum limbs. Dev Biol. 2016;416:26–33.
Satoh A, Mitogawa K, Makanae A. Regeneration inducers in limb regeneration. Develop Growth Differ. 2015;57:421–9.
Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–8.
Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. Cold Spring Harbor Lab. 2011;3:a004952.
Scott JE, Dis JDAR. Differential staining of acid glycosaminoglycans (mucopolysaccharides) by alcian blue in salt solutions. His-tochemie. 1965;5:221–33 Fletcher CDM ….
Ochoa MT, Loncaric A, Krutzik SR, Becker TC, Modlin RL. "dermal dendritic cells" comprise two distinct populations: CD1+ dendritic cells and CD209+ macrophages. J Invest Dermatol. 2008;128:2225–31.
Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci U S A. 2013;110:9415–20.
Marrero L, Simkin J, Sammarco M, Muneoka K. Fibroblast reticular cells engineer a blastema extracellular network during digit tip regeneration in mice. Regeneration (Oxf). 2017;4:69–84.
Neufeld DA, Day FA. Perspective: a suggested role for basement membrane structures during newt limb regeneration. Anat Rec. 1996;246:155–61.
McCusker CD, Athippozhy A, Diaz-Castillo C, Fowlkes C, Gardiner DM, Voss SR. Positional plasticity in regenerating Amybstoma mexicanum limbs is associated with cell proliferation and pathways of cellular differentiation. BMC Dev Biol BioMed Central. 2015;15:45.
Bodeen WJ, Marada S, Truong A, Ogden SK. A fixation method to preserve cultured cell cytonemes facilitates mechanistic interrogation of morphogen transport. Development. 2017;144:3612–24.
Ramírez-Weber FA, Kornberg TB. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 1999;97:599–607.
González-Méndez L, Gradilla A-C, Guerrero I. The cytoneme connection: direct long-distance signal transfer during development. Development. 2019;146:dev174607.
Krystel-Whittemore M, Dileepan NK, Wood JG. Mast cell: a multi-functional master cell. Front Immunol. 2015;6:620.
Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, Liu H, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22:303–7.
Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119.
Gardiner DM, Muneoka K, Bryant SV. The migration of dermal cells during blastema formation in axolotls. Dev Biol. 1986;118:488–93.
Tank PW, Connelly TG, Bookstein FL. Cellular behavior in the anteroposterior axis of the regenerating forelimb of the axolotl, Ambystoma mexicanum. Dev Biol. 1985;109:215–23.
Iten LE, Bryant SV. The interaction between the blastema and stump in the establishment of the anterior--posterior and proximal--distal organization of the limb regenerate. Dev Biol. 1975;44:119–47.
Stocum DL. The urodele limb regeneration blastema: a self-organizing system. i. Differentiation in vitro. Dev Biol. 1968;18:441–56.
French V, Bryant PJ, Bryant SV. Pattern regulation in epimorphic fields. Science. 1976;193:969–81.
Bryant SV, French V, Bryant PJ. Distal regeneration and symmetry. Science. 1981;212:993–1002.
Kornberg TB, Roy S. Cytonemes as specialized signaling filopodia. Development. Oxford University Press for The Company of Biologists Limited; 2014;141:729–736.
Eom DS, Parichy DM. A macrophage relay for long-distance signaling during postembryonic tissue remodeling. Science. American Association for the Advancement of Science. 2017;355:1317–20.
Eom DS, Bain EJ, Patterson LB, Grout ME, Parichy DM. Long-distance communication by specialized cellular projections during pigment pattern development and evolution. Elife. eLife Sciences Publications Limited. 2015;4:2686.
Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–21.
Poulain FE, Yost HJ. Heparan sulfate proteoglycans: a sugar code for vertebrate development? Development. 2015;142:3456–67.
Bülow HE, Berry KL, Topper LH, Peles E, Hobert O. Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc Natl Acad Sci U S A. 2002;99:6346–51.
Laurencin CT, Khan Y. Regenerative Engineering. Boca Raton: CRC Press, Taylor & Frances Group; 2013.
Acknowledgements
We wish to thank the members of the Bryant/Gardiner Lab for help with and encouragement of the research. Mouse limb skin tissue samples were kindly provided by Dr. David Rowe. Research was supported by an NIH Director’s Pioneer Award, and the National Science Foundation through its support of the Ambystoma Genetic Stock Center at the University of Kentucky, Lexington. Dr. Laurencin was the recipient of the Presidential Faculty Fellow Award from the National Science Foundation.
Author information
Authors and Affiliations
Contributions
SB and DG conceptualized the presence of a positional information GRID; DG, AP, and TO designed the experiments; AP and TO conducted the experiments; DG, AP and TO wrote the draft of the manuscript; and all authors contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
None of the authors have competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
T. Otsuka and A. Q. Phan are co-first authors.
Electronic supplementary material
Supplementary Figure 1
Visualization of gels from RT-PCR analysis of expression of axolotl gene that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in axolotl limb skin, as well as blastema tissue during limb regeneration. (PDF 5239 kb)
Supplementary Figure 2
Visualization of gels from RT-PCR analysis of expression of axolotl gene that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in a diversity of tissues. (PDF 11416 kb)
Supplementary Table 1
Results from a search of the axolotl EST database (ref) for genes expected to be involved in both the synthesis of GAG chains and in modification of their patters of sulfation. PCR primers were synthesized as indicated for RT-PCR analysis of expression (Supplemental Fig. 1a, b, and Supplemental Table 2). (PDF 2259 kb)
Supplemental Table 2
Summary of the relative levels of expression of axolotl genes (RT-PCR analysis illustrated in Supplemental Fig. 1a, b) that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in a diversity of tissues, as well as during limb regeneration. (PDF 57 kb)
Rights and permissions
About this article
Cite this article
Otsuka, T., Phan, A.Q., Laurencin, C.T. et al. Identification of Heparan-Sulfate Rich Cells in the Loose Connective Tissues of the Axolotl (Ambystoma mexicanum) with the Potential to Mediate Growth Factor Signaling during Regeneration. Regen. Eng. Transl. Med. 6, 7–17 (2020). https://doi.org/10.1007/s40883-019-00140-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40883-019-00140-3