Abstract
The ability to precisely modify genomic sequences has always been a powerful tool for determining the relationship between genotype and phenotype, and for targeted editing of disease-causing genetic lesions. Until recently, it has been difficult to routinely achieve this goal. The advent of CRISPR/Cas9 has resulted in the widespread adoption of genome editing as a standard laboratory procedure and has accelerated the options available for researchers and clinicians working on accurate in vitro cell models of disease, or for the development of future ex vivo and in vivo gene therapies. In this review, we provide an update on CRISPR/Cas9 technologies and delve into how this tool can be used for stem cell engineering, focusing on the most pressing and feasible applications. Lastly, we briefly discuss the ethical concerns that have caused worldwide alarm in the stem cell field. Despite the technical challenges that still lie ahead, CRISPR-based genome editing offers unprecedented opportunities for advancing stem cell therapies to the clinic.
Similar content being viewed by others
Introduction
In November 2015, a collaboration of clinicians and scientists at Great Ormond Street Hospital in London infused T lymphocytes engineered to express chimeric antigen receptors (CARs) into a 1-year-old infant with refractory relapsed B cell acute lymphoid leukaemia. Astonishingly, allogeneic (mismatched) donor T cells were used for the infusion. The breakthrough involved the application of gene-editing tools to modify the HLA genes in donor cells in order to eliminate the risk of graft-versus-host disease (GvHD) and provide a readily available supply of therapeutic T lymphocytes. In this single example of compassionate use therapy, researchers were able to report no trace of leukaemia and attributed the success to protected engineered donor cells [1]. This highlighted example represents the power of gene editing to propel a biomedical revolution and meet the challenges of potentially curing genetic diseases. While genome editing tools have been available for over 20 years, with some having made their way to the clinic, it is only recently that genome engineering has become a facile and readily available option for therapeutic use. The protagonist is a technology called CRISPR/Cas9 or clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) protein 9. CRISPR/Cas9 has brought the promise of gene editing as a therapy a lot closer to reality. Few new areas of biology have roused quite the same level of enthusiasm (and also ethical concern). CRISPR/Cas9 will no doubt have a profound and far-reaching effect on the field of stem cell research and therapy. Here, we describe this burgeoning genome editing phenomenon and discuss the most likely stem cell applications to be early beneficiaries for clinical implementation. Lastly, we touch on the ethical concerns, which have brought unprecedented levels of public attention to the therapeutic stem cell field.
The Basics of Gene Editing
The gene editing toolbox encompasses several types of molecular scissors. Zinc finger nucleases (ZFN) were one of the first editing tools to show specific targeting of the genome as exemplified nearly 25 years ago in Drosophila [2]. More recently, transcription activator-like effector nucleases (TALENs) derived from plant pathogens were discovered and applied to gene-editing approaches [3]. These were shown to be simpler to design and less expensive than ZFNs but still required significant expertise and resources. CRISPR/Cas9 is derived originally from an endogenous bacterial defence mechanism [4, 5]. In its most simplified form, it functions with two modified components: a short chimeric single guide RNA (sgRNA) and a Cas9 nuclease [6••]. The sgRNA acts as homing guide for Cas9, thus making it easy to target different genomic loci by simply replacing the sgRNA sequence in the presence of the same nuclease (Fig. 1). It is this flexibility and ease of use which has allowed CRISPR/Cas9 to be adopted widely, where it has been used to target DNA sequences from a number of different organisms [7••, 8••, 9••]. CRISPR/Cas9 has been described by one of the pioneers of gene editing, George Church, as ‘a real gift from biology’ [10].
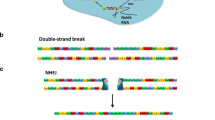
Molecular manipulations and stem cell applications of CRISPR/Cas9. a Classic CRISPR/Cas9 design leads to small guide RNA (sgRNA) sequence-specific targeting of the CRISPR/Cas9 nuclease to a desired sequence, leading to a double strand break. The break is then rapidly repaired by the cell using either NHEJ or HDR. The former is an error-prone pathway, which yields random insertions or deletions (indels) modifying the original sequence. These can be targeted within genes to create frameshift mutations leading to knockout of functional gene product. Alternatively, a donor template with homology to the surrounding sequence can be provided with the CRISPR/Cas9 to lead to a specific and desired change. The applications of this type of modification show enormous promise for the ex vivo modification of patient-derived cells to correct disease-associated mutations for cell replacement therapy. Isogenic models of disease can also be generated with relative ease using this model. b, c show modifications of CRISPR/Cas9 in the form of deactivated Cas9 (dCas9), removing the nuclease capability of Cas9. b dCas9 can recruit transcriptional modifiers to specific DNA sequences. Genes can therefore be activated or repressed at their endogenous loci, and further epigenetically modified at functional DNA elements. Such molecular manipulations can be exploited to induce pluripotency and investigate transcriptional regulation in lineage commitment and the mechanisms of enhancer elements. c dCas9 can also be tweaked to recruit other molecules including functional RNAs (e.g., aptamers and long noncoding RNAs [lncRNAs] bound to effector proteins). This technique shows great potential as a tool to investigate the role of lncRNAs at specific DNA sequences in transcriptional control
The CRISPR/Cas9 system is malleable and can be used for applications beyond merely editing. The Cas9 nuclease can be deactivated (‘dead’ or dCas9), allowing for an array of applications that require programmable RNA-dependent DNA-binding proteins [11, 12•]. dCas9 fused to accessory proteins, such as transcription activator and repressor domains, provide RNA-guided targeted interactions at specific loci [11, 12•, 13–16]. Unlike TALENS and ZFNs, CRISPR/Cas9 technologies have the advantage of being ‘programmable’, owing to the sgRNA being physically separated from the nuclease domain. This allows for the potential to target multiple sites simultaneously, facilitating large-scale genomic editing.
Although each type of gene-editing technology uses a different mechanism to recognise a specific sequence DNA, all create a nuclease-dependent double strand break in the DNA. The cellular machinery then rapidly responds to repair the damage using one of two mechanisms: the cell (1) can use its ‘quick-fix’ nonhomologous end joining (NHEJ) mechanism to seal the break or (2) use the specific homologous DNA repair (HDR) machinery in conjunction with a DNA template to generate a desired genetic change. In the first case, often random insertions or deletions (in/dels) are generated; thus if a break is generated in the correct place, one could rely on incorrect in/dels to lead to a ‘broken’ unreadable gene, thus yielding a knockout of gene function. In the second, a modification can be made to introduce a desired genetic change at the targeted genomic region, which can range from changing a single nucleotide to incorporating an entirely different sequence [17].
Cas9-catalysed DNA cleavage is guided by a 17–20 nucleotide sgRNA, resulting in deletions generated by NHEJ at the binding site, or alternatively, in HDR (Fig. 1a). Traditionally, NHEJ has been painfully inefficient, particularly in human stem cells. The advent of the CRISPR/Cas9 revolution has challenged this notion, with, in some cases, a log fold improvement in editing efficiencies of CRISPR/Cas9 over TALENs [18, 19]. However, with this substantial increase in efficiency comes a greater propensity for off-target effects (recently and comprehensively reviewed by Zhang et al. [20]). Thus, from a therapeutic point of view, CRISPR/Cas9 tools have to be substantially improved before clinical implementation.
Modifications of CRISPR/Cas9
The aforementioned dCas9 can also be used to recruit transcriptional modulators at specific sites to investigate stem cell biology at the regulatory level (Fig. 1b). The recruitment of transcriptional coactivators such as VP64 has been exploited to activate stem cell master regulators to induce pluripotency [21] as well as to direct mouse embryonic fibroblasts towards skeletal muscle [22]. Other derivatives such as that fused to the acetyltransferase p300 have been shown to switch on pluripotency genes in HEK293 cell lines by directly modifying the epigenetic marks at the endogenous locus [23]. There are, in addition, numerous other modifications of Cas9 that have been shown to be valuable molecular tools. One that deserves mention is CRISPR-Display, in which the sgRNA is modified to include large functional RNA sequences depositing them at specific sites [24•]. The roles of functional noncoding RNAs in regulating cellular reprogramming [25] and lineage specification (recently reviewed Rosa and Ballarino, and Hu et al. [26, 27]) are becoming of great interest, particularly in our understanding of epigenomic plasticity in development (Fig. 1c). Although the application of CRISPR-Display at endogenous loci may need to overcome some technical hurdles, it shows great promise in providing a desperately needed tool to distinguish the roles of a functional DNA element from its encoded noncoding RNA transcript.
In vitro Stem Cell Models of Disease
Applications of CRISPR/Cas9 range from generating isogenic models of disease to providing a malleable molecular tool with which to interrogate and modulate the genome for basic biological research. Many of these have been exquisitely reviewed by Sternberg and Doudna [28]; therefore, we will limit ourselves to those with direct stem cell applications.
Stem cell models of disease have proved, of their own accord, to be a powerful tool in elucidating pathological mechanisms in vitro [29], not least of which includes the use of induced pluripotent stem cells (iPSCs) [30]. These cells retain the genotype of the individual from which they are derived and, in addition, can be manipulated to differentiate into almost any cell type in the body, making them a valuable tool in uncovering the genetic sources of disease. However, such genetic components could be confounded by the background genetic variation among individuals. Indeed, it was only upon the generation of an isogenic iPSC-derived cellular model of Parkinson’s disease through gene editing that researchers were able to extract differential transcriptional changes caused by a missense mutation in the α-synuclein gene [31•]. Thus, the generation of isogenic stem cell models of disease is becoming more commonplace [32]. Isogenic in vitro models can also facilitate preclinical work, as Li and colleagues recently showed in a CRISPR-generated iPSC-derived model of Duchenne Muscular Dystrophy (DMD) [33]. The authors were able to identify unique regions and assess off-target effects at a genome-wide scale. The study further investigated the efficacy of three different gene-editing strategies including repairing a splice acceptor site, introducing a frameshift mutation to exclude a disrupted exon and, finally, the introduction of the missing exon. Subsequent differentiation of repaired isogenic clones into skeletal muscle showed restored levels of the dystrophin protein. Another CRISPR-based study repaired an inversion mutation in haemophilia patient-derived iPSCs [34]. Importantly, this report demonstrated that differentiated endothelial cells from repaired iPSCs were able to functionally rescue lethality in a mouse model of haemophilia [34].
Most recently, CRISPR/Cas9 was used to correct a missense mutation in the chronic granulomatous disease (CGD)-causing gene, CBB, in patient-derived iPSCs [35]. This study further emphasises the advantages of using isogenic models in that it showed a restoration of reactive oxygen species (ROS) production after correcting the mutation. Therefore, gene editing in the sphere of in vitro stem cell modelling can not only help us elucidate novel disease mechanisms but also provide valuable preclinical data to support its use therapeutically.
Although CRISPR/Cas9 stem cell models are now designed with relative ease, there is a great need to improve the methodology for low efficiency modifications. These include strategies in which large sequences need to be incorporated into the genome from which clones are to be derived or screening applications where a high gene-editing efficiency is necessary across the global cell population without subsequent clonal selection. However, the efficiency of site-specific modifications is poorest in models that would provide the most physiological relevance, i.e., pluripotent and haematopoietic stem cells and their cellular derivatives. This is due in no small part to their refraction to plasmid transfection, although this can be dramatically improved through the use of protein/RNA complexes [36•].
One intriguing approach used to circumvent the problem of editing efficiencies was the report of the induced CRISPR (iCRISPR) system in which TALENs were used to incorporate an inducible Cas9 expression cassette within the AAVS1 safe harbour locus [37]. Importantly, this locus has been extensively identified as a region into which genomic integration has no observable detrimental effects [38–40]. The use of iCRISPR could greatly enhance the CRISPR armoury since it leads to Cas9 expression only upon doxycycline exposure, yielding inducible Cas9 expression. PSCs harbouring this iCRISPR system can be directed, at any point, to produce Cas9. Simultaneously, sgRNAs can be transfected and are readily taken up by otherwise traditionally difficult to transfect stem cell derivatives. One could envisage a highly effective and malleable system in which gene editing could be induced at different time points in lineage specification, providing a powerful tool for research into cell fate and determination. Other reports have demonstrated alternatives for providing inducible CRISPR/Cas9 through the use of optogenetics [41] and the destabilisation domain-tagged essential protein approach [42]. These, in combination with additional studies, have incorporated the use of small molecules to either synchronise cells to increase HDR efficiency [43] or used compound screens to identify drugs that appear to enhance both HDR or NHEJ [44•]. This should provide further enhancement of the CRISPR/Cas9 toolbox in stem cells.
Organoid Models
While the monolayer cultures discussed above have been invaluable, there is an increasing appreciation to more closely mimic the cellular niche. Organoids are stem cell-derived three-dimensional structures generated in vitro and have been made to mimic several organs in the body ranging from mini-gut to mini-brain [45]. Importantly, they offer the advantage over monolayer stem cell models of replicating some 3D physiological processes and responses only seen in vivo, perhaps best shown in the intestinal stem cell-based ‘mini-gut’ models developed by Hans Clevers [46•]. Because of this physiological relevance, these models are likely to shorten the drug pipeline significantly. Indeed, gene editing of organoids has already been demonstrated extremely valuable in several examples. In 2013, Hans Clevers’ group showed that mini-gut organoids derived from cystic fibrosis (CF) patients could be genetically corrected using CRISPR/Cas9 [47•]. In this first proof-of-concept study, the authors showed that only corrected organoids were able to respond to increased cAMP levels, one of the critical disease phenotypes observed in the intestines of CF patients. More recently, Matano and colleagues showed how CRISPR/Cas9 could be used in the sphere of colorectal cancer research [48]. The report demonstrated that the addition of known mutations into organoids derived from normal human intestinal epithelium was not sufficient to induce macrometastases, but that such engineered mutations were only effective within the background of a chromosomally unstable adenoma-derived organoid. The discovery of the contributing factors to the invasive nature of tumours in this study demonstrates that combining gene editing with organoid modelling may be more than critical in elucidating disease mechanisms in stem cell-derived models.
Stem Cell Screening Using CRISPR/Cas9
For the most part, stem cell models, whether monolayer or 3D are generated with the purpose of studying a few changes to the genome. In contrast, the development of CRISPR/Cas9 libraries used to target genes at a global level has already provided extensive data on the roles of genes essential to several biological processes including infectious agents, cancer and pluripotency [49–53]. Such screens may be applied to several avenues of stem cell research ranging from the identification of novel drug targets to the enhancement of ex vivo bulking of haematopoietic stem cells. CRISPR/Cas9-based screens offer the advantage of assaying the effect of complete loss of gene expression compared to a reduction in gene expression when using gene knockdown screens. Moreover, we now have the ability to directly interrogate the function of noncoding DNA elements in their role as key players in stem cell nuclear architecture [54]. A classic example of this was shown by the loss of pluripotency in mouse embryonic stem cells (mESC) after the discreet removal of the mouse Sox2 super enhancer element 100 kb downstream of the Sox2 locus [55•]. Indeed, intriguing evidence from such screens suggests that the CRISPR/Cas9-mediated disruption of noncoding elements can lead to a highly cell-type-specific lethal phenotype [51]. Application of such screens to the investigation of contributing effects of chromosomal architecture in lineage commitment is likely to significantly improve our understanding of stem cell differentiation.
Direct Therapeutic Strategies of CRISPR/Cas9 in Stem Cells
While the mechanistic applications of CRISPR/Cas9 reveal the powerful way in which this system can be exploited to advance our understanding of basic biology, it is the therapeutic application of this technology that has caught the public’s attention. Today, a plethora of examples of successful gene-editing approaches exists [56].
There are several strategies in which gene editing can be applied for direct cell or gene therapies, once a genetic target has been identified: ex vivo, progenitor cells from a patient can be removed, repaired and subsequently replaced in the same individual; germ line/zygote correction, cells that hold genetic material carried throughout the entire organism can be modified leading to a permanent change in subsequent generations; and in vivo, modification of the DNA in cells in a patient followed by direct addition of the CRISPR/Cas9 tool itself.
Editing ex vivo for Cell Replacement Therapy
To date, the ex vivo gene-editing approach in humans is the most advanced strategy of those listed above. This is largely due to the extensive prior art of expanding haematopoietic stem cells for the treatment of blood disorders. In 2014, Xie and colleagues performed a proof-of-concept study where they used CRISPR/Cas9 to correct mutations in the HBB gene, which causes β-thalassemia [57]. Repaired patient-derived iPSC clones were then expanded into red blood cell precursors in vitro and shown to have increased levels of HBB expression compared to uncorrected cells.
Interestingly, it is likely that the first successful application of ex vivo gene editing will come through the targeting a human gene required for HIV infection: CCR5. It is well established that individuals homozygous for a relatively common Δ32 mutation in the CCR5 gene are able to control levels of R5 tropic viral replication to the extent that they are clinically asymptomatic even without treatment [58]. Indeed, in 2008, a ground-breaking procedure of replacing a patient’s entire haematopoietic system with CCR5-null bone marrow led to the only patient to date to have been clinically ‘cured’ of AIDS [59]. Thus, editing the CCR5 gene in CD4+ T cell is one of the leading ex vivo strategies and has been expanded from a phase I to II clinical trial [60•, 61]. CRISPR/Cas9-mediated gene editing has recently been used to target the same gene in two studies: first in macrophages derived from human iPSCs subsequently shown to resist in vitro HIV infection [62] and, secondly, in haematopoietic CD34+ progenitors [63]. In the context of HIV treatment, it is worth noting that CRISPR/Cas9 has further been used to target the alternative HIV coreceptor, CXCR4 in primary CD4+ T cells, leading to inhibition of HIV infection [64]. Since the prolonged expansion of T cells ex vivo can lead to detrimental cellular and genetic effects, increasing the efficiency of editing ex vivo remains critical. As described earlier, another blood disease for which CRISPR/Cas9 has been successfully applied is CGD [35]. This study not only provided valuable information from a modelling perspective but also importantly demonstrated that clinically relevant efficiencies of editing using CRISPR/Cas9 are attainable, which would greatly facilitate an ex vivo treatment without requiring extensive and potentially damaging passaging of haematopoietic cells. In cases where a threshold level of repair cannot be achieved, it is worth noting that chemical modifications enhancing the stability of sgRNAs in conjunction with an enhanced Cas9/sgRNA delivery method have been shown to dramatically increase editing in CD34+ haematopoietic progenitors in both stimulated and unstimulated T cells [65]. These modifications are therefore likely to have an important impact on haematopoietic stem cell therapies.
While haematopoietic disorders have been used as the driving force to test genome editing, other cell types have also been targeted. Perhaps one of the most intriguing approaches is to combine the efficiency of CRISPR/Cas9 editing with 3D organoids for therapeutic use. We have already discussed the development of gene-edited organoids as a molecular tool; however, such models may have great potential in direct therapeutic application. Indeed, in vitro generated gut organoids have been shown to be successfully transplanted in vivo resulting in phenotypic improvement in the damaged colons of mice [66]. Thus, an ideal strategy to alleviate the clinical symptoms associated with the intestinal phenotype of CF in humans may be to repair the mutation ex vivo (as was shown by Schwank and colleagues [47•]) and generate gut organoids (or lung stem cells) to replace the defective intestinal layer. Other organoids like the mini-liver [67] also show great promise for replacing damaged organs. It is likely that proof-of-principle studies such as these may pave the way towards direct therapeutic intervention with gene-corrected organoids.
Editing the Germ Line and Ethical Considerations
While editing a deleterious mutation ex vivo leads to a repair in only a portion of an individuals’ cells, editing at the one cell embryo stage allows for a permanent DNA alteration inheritable to subsequent generations. An exhaustive number of studies have shown the efficacy of CRISPR/Cas9-mediated gene editing in the germ lines of species ranging from worms to primates (reviewed by Rocha-Martins et al. [68]). CRISPR/Cas9 offers the advantage over other gene-editing strategies of being easily multiplexed, thus negating the labour and time traditionally associated with transgenic animal generation [69•].
The success this system has achieved in transgenic animal modelling (particularly in nonhuman primates [70]) naturally begs the question of whether the same could be done in human embryos harbouring inherited disease mutations. Proof-of-principle studies in mice have shown that both cataracts [71] and DMD [72] can be effectively cured in offspring after CRISPR/Cas9-mediated repair of the disease-associated mutations. More recently, Reddy and colleagues demonstrated that human mitochondrial DNA harbouring deleterious mutations causing mitochondrial diseases could be reduced after gene editing in mouse oocytes [73].
However, only one study to date has reported editing in human embryos in which the effects of CRISPR/Cas9 editing targeting the HBB gene in tripronuclear human zygotes was assessed [74••]. The results showed limited success in modifying the genome. The authors raised a number of concerns regarding the ‘readiness’ of the application of CRISPR/Cas9 editing based on their results, but two were cause for alarm: (1) The few modified embryos were genetically mosaic, and (2) A high degree of off-target effects was observed.
The propensity with which CRISPR/Cas9 has revolutionised the arena of gene editing has led to almost unparalleled public interest and created a highly polarised debate, particularly in light of the human embryo editing report [74••, 75]. This interest led to an extraordinary gathering of international scientists at the National Academy of Sciences in Washington D.C. in late 2015, to discuss a potential moratorium on human embryo editing [76]. Interestingly, a blanket moratorium was not endorsed, but rather, the panel argued for strict regulatory body-approved applications.
The most notable concern is that of the generation of ‘designer babies’. However, there remain viable technical reasons why this technology is not ready for embryo modification, and most centre on eliminating or selecting-out deleterious off-targets effects. Another factor that negates the use of edited embryos lies in the fact that few known genetic conditions exist for which preimplantation genetic diagnosis (PGD) cannot be used to screen-out embryos with pathogenic mutations. Moreover, three parent babies (in the case of mitochondrial disease) cannot be used to exclude inheritance of disease-associated mutations. Nevertheless, as recently commented by Paul Knoepfler, patients carrying incurable mutations are likely to have a higher degree of risk tolerance than scientists [77].
Concluding Remarks
CRISPR/Cas9 may well be considered one of the most important biological tools identified in recent years. There is no end to the number of ways this system can be tweaked for a vast array of molecular exploits. This is especially the case in stem cells, where the application of CRISPR/Cas9 technologies will likely be profound (and also heavily debated). Although hurdles remain, especially the warranted concern of off-target effects, continued improvements in Cas9 and guide RNA engineering will allay the fears of even the most ardent critics of CRISPR-based therapy [78]. Already, there is unprecedented investment by large and small biotechnology and pharmaceutical industry players alike, which shows the palpable excitement generated by the vast array of possibilities brought by this technology. Eventually, the early hype will subside, but there is little doubt that CRISPR/Cas9, and its application in stem cell engineering, will have an extraordinary impact in the progress towards curing many previously intractable diseases.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Paper: Multiplex Cripsr/Cas9 Genome Editing to Generate Potent Universal CART and PD1-Deficient Cells Against Leukemia. [Online]. Available: https://ash.confex.com/ash/2015/webprogram/Paper83406.html. [Accessed: 19-Jan-2016].
Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161(3):1169–75.
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8.
Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–82.
Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–40.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. This report was the first to demonstrate that CRISPR/Cas9 could be used to create a break in a sequence specific manner, and therefore, the first to demonstrate that it could be used as a gene editing tool.
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2, e00471. Published in quick succession in 2013, and showed that the CRISPR/Cas9 system could be used to edit the genome in cells. Cong et al. were the first to show editing in stem cells.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9’. Science. 2013;339(6121):823–6. Published in quick succession in 2013, and showed that the CRISPR/Cas9 system could be used to edit the genome in cells. Cong et al. were the first to show editing in stem cells.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. Published in quick succession in 2013, and showed that the CRISPR/Cas9 system could be used to edit the genome in cells. Cong et al. were the first to show editing in stem cells.
Genome Editing with CRISPRs, TALENs and ZFNs | Biocompare: The Buyer’s Guide for Life Scientists. [Online]. Available: http://www.biocompare.com/Editorial-Articles/144186-Genome-Editing-with-CRISPRs-TALENs-and-ZFNs/. [Accessed: 19-Jan-2016].
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–51.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–83. This was one of the earliest papers, which was subsequently validated by a number of other publications, to show how CRISPR/Cas9 could be deactivated to remove the nuclease activity and tweaked to recruit transcriptional modifiers to specific sites.
Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23(10):1163–71.
Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10(10):977–9.
Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10(10):973–6.
Hu J, Lei Y, Wong W-K, Liu S, Lee K-C, He X, et al. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic Acids Res. 2014;42(7):4375–90.
Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26(1):52–64.
Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41(19):9049–61.
Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12(4):393–4.
Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4, e264.
Gao X, Yang J, Tsang JCH, Ooi J, Wu D, Liu P. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem Cell Rep. 2013;1(2):183–97.
Chakraborty S, Ji H, Kabadi AM, Gersbach CA, Christoforou N, Leong KW. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Rep. 2014;3(6):940–7.
Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–7.
Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12(7):664–70. This report showed how the CRISPR/Cas9 system could be tailored to recruit functional RNAs to specific sites in the genome.
Kim DH, Marinov GK, Pepke S, Singer ZS, He P, Williams B, et al. Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell Stem Cell. 2015;16(1):88–101.
Rosa A, Ballarino M. Long noncoding RNA regulation of pluripotency. Stem Cells Int. 2016;2016:1797692.
Hu S, Shan G, Hu S, Shan G. LncRNAs in stem cells, LncRNAs in stem cells. Stem Cells Int. 2016;2016(2016), e2681925.
Sternberg SH, Doudna JA. Expanding the Biologist’s toolkit with CRISPR-Cas9’. Mol Cell. 2015;58(4):568–74.
Sterneckert JL, Reinhardt P, Schöler HR. Investigating human disease using stem cell models. Nat Rev Genet. 2014;15(9):625–39.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72.
Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146(2):318–31. This is the first report to demonstrate within the context of iPSC-derived models, that isogenic cell lines generated by editing could be incredibly informative in uncovering phenotypic changes associated with single gene mutations by removing the complicating background genetics between different patient derived cell lines.
Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech. 2013;6(4):896–904.
Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9’. Stem Cell Rep. 2015;4(1):143–54.
Park C-Y, Kim DH, Son JS, Sung JJ, Lee J, Bae S, et al. Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9’. Cell Stem Cell. 2015;17(2):213–20.
Flynn R, Grundmann A, Renz P, Hänseler W, James WS, Cowley SA, et al. CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Exp Hematol. 2015;43(10):838–48. e3.
Kim S, Kim D, Cho SW, Kim J, Kim J-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–9. This report demonstrated the extent to which inefficient genome editing in stem cells could be significantly improved by reducing the CRISPR/Cas9 plasmid to an RNP complex.
González F, Zhu Z, Shi Z-D, Lelli K, Verma N, Li QV, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15(2):215–26.
Tiyaboonchai A, Mac H, Shamsedeen R, Mills JA, Kishore S, French DL, et al. Utilization of the AAVS1 safe harbor locus for hematopoietic specific transgene expression and gene knockdown in human ES cells. Stem Cell Res. 2014;12(3):630–7.
DeKelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20(8):1133–42.
Kotin RM, Linden RM, Berns KI. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11(13):5071–8.
Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11(3):198–200.
Park A, Won ST, Pentecost M, Bartkowski W, Lee B. CRISPR/Cas9 allows efficient and complete knock-in of a destabilization domain-tagged essential protein in a human cell line, allowing rapid knockdown of protein function. PLoS One. 2014;9(4), e95101.
Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3, e04766.
Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–7. References 43 and 44 showed how editing in stem cells could be improved by the use of synchronization and small molecules.
Orqueda A, Javier S, Gimenez CA, Pereyra-Bonnet F. iPSCs: a minireview from bench to bed, including organoids and the CRISPR system. Stem Cells Int. 2016;2016, e5934782.
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. This paper was the first to demonstrate the development of an in vitro organoid model in the form of a mini-gut.
Schwank G, Koo B-K, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–8. Here Hans Clevers' group used CRISPR/Cas9 to repair mutations in minigut organoid models derived from cystic fibrosis patients, showing for the first time the combined power of organoids with CRISPR/Cas9 for potential therapeutic intervention.
Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21(3):256–62.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–7.
Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–4.
Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350(6264):1096–101.
Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–91.
Toledo CM, Ding Y, Hoellerbauer P, Davis RJ, Basom R, Girard EJ, et al. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Rep. 2015;13(11):2425–39.
Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–6. Bing Ren and colleagues showed the extensive chromatin reorganization that occurs through loss of pluripotency and through lineage specification.
Li Y, Rivera CM, Ishii H, Jin F, Selvaraj S, Lee AY, et al. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014;9(12), e114485. In this report the authors show an exquisitely simple manner in which CRISPR/Cas9 can be used as a molecular tool to uncover the contributions of a distal super enhancer to gene regulation of one of the key pluripotency genes, Sox2.
Maeder ML, Gersbach CA. Genome editing technologies for gene and cell therapy. Mol Ther J Am Soc Gene Ther. 2016.
Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, et al. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24(9):1526–33.
Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–5.
Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–8.
Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–10. At present this study represents the most advanced clinical trial published using ex vivo gene-edited cells for cell therapy.
Sangamo BioSciences Presents Phase 2 Clinical Data From Two SB-728-T HIV Studies (NASDAQ:SGMO). [Online]. Available: http://investor.sangamo.com/releasedetail.cfm?ReleaseID=946687. [Accessed: 19-Jan-2016].
Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci U S A. 2014;111(26):9591–6.
Mandal PK, Ferreira LMR, Collins R, Meissner TB, Boutwell CL, Friesen M, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9’. Cell Stem Cell. 2014;15(5):643–52.
Hou P, Chen S, Wang S, Yu X, Chen Y, Jiang M, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci Rep. 2015;5:15577.
Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33(9):985–9.
Xue X, Shah YM. In vitro organoid culture of primary mouse colon tumors. J Vis Exp. 2013;75.
Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499(7459):481–4.
Rocha-Martins M, Cavalheiro GR, Matos-Rodrigues GE, Martins RAP. From gene targeting to genome editing: transgenic animals applications and beyond. An Acad Bras Cienc. 2015;87(2 Suppl):1323–48.
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. This represents one of the first reports to demonstrate the ease with which CRISPR/Cas9 could be exploited to generate transgenic mice.
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–43.
Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9’. Cell Stem Cell. 2013;13(6):659–62.
Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345(6201):1184–8.
Reddy P, Ocampo A, Suzuki K, Luo J, Bacman SR, Williams SL, et al. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell. 2015;161(3):459–69.
Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–72. To date this remains the only published study to have experimented with CRISPR/Cas9 in human zygotes.
Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348(6230):36–8.
International Summit on Human Gene Editing. [Online]. Available: http://www.nationalacademies.org/gene-editing/Gene-Edit-Summit/index.htm. [Accessed: 20-Jan-2016].
admin, ‘Just freakin do it: patients voice impatience on CRISPR for genetic diseases’, The Niche, 15-Dec-2015. [Online]. Available: https://www.ipscell.com/2015/12/just-freakin-do-it-patients-voice-impatience-on-human-gene-editing/. [Accessed: 20-Jan-2016].
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–8.
Acknowledgments
Research reported in this publication was supported by the Strategic Health Innovation Partnerships (SHIP) Unit of the South African Medical Research Council with funds received from the South African Department of Science and Technology. Funding from the National Research Foundation is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Janine Scholefield and Marc S. Weinberg declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Genome Editing
Rights and permissions
About this article
Cite this article
Scholefield, J., Weinberg, M.S. The Application of CRISPR/Cas9 Technologies and Therapies in Stem Cells. Curr Stem Cell Rep 2, 95–103 (2016). https://doi.org/10.1007/s40778-016-0043-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-016-0043-7