Abstract
Purpose of Review
Candida auris is a multi-drug-resistant pathogen with many phenotypic variations that contribute to its pathogenicity. This review aims to characterize its phenotypic heterogeneity while highlighting the variants that should be prioritized in future research to advance therapies against C. auris.
Recent Findings
As the Earth warms, fungi like Candida experience selective pressure to tolerate these higher temperatures and become the few fungal species capable of successfully colonizing the host. The most recent of these is C. auris, which has become an acute concern due to its rapid emergence, high mortality rate, and resistance to all known classes of antifungals.
Summary
Several studies have contributed rapidly to our general understanding of C. auris, but not enough has been experimentally verified on its morphological variation and its ability to maintain a successful commensal lifestyle on the human skin. Because of its distinct phenotypic variations compared to other fungal species, especially under the selective pressures of its host, C. auris presents a unique opportunity to identify unique targets and strategies to contribute to the antifungal pipeline and control emergent pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 2019, Candida auris became the first fungal pathogen to be declared a global health threat by the CDC [1]. Though the more prevalent species, Candida albicans, is to date the most common causative agent of candidemia in the USA [2], C. auris represents an acute concern due to its rapid emergence, high mortality rate, and resistance to all three classes of antifungals [3]. Treatment of C. auris is further complicated by its high transmissibility within intensive care and hospital units. C. auris has been found to linger on hospital surfaces and equipment, remaining resistant to desiccation and disinfection for at least several weeks [4, 5]. In the wake of the COVID-19 pandemic, which overflowed hospitals and ICUs, C. auris experienced markedly rising incidence and mortality rates in the USA, presumably due to the increasing number of immunocompromised patients in hospitals, ICUs, and nursing homes [6].
C. auris is one of many fungal infections that plague several other organisms, but fungal intolerance of high temperatures has protected most endotherms, including humans, from these infections. However, as the Earth’s climate warms due to anthropogenic climate change, fungi across ecosystems experience selective pressure to tolerate these higher temperatures. The Candida genus, including C. auris, which can tolerate temperatures greater than 40 °C, are among the few fungal species capable of colonizing the human host. As such, they represent a unique opportunity to identify novel targets and strategies for treating fungal infections. This is of crucial importance, as fungi represent a major reservoir of possible novel human pathogens. The threat only grows as we surpass 1 °C of global temperature increase since the pre-industrial age [7].
In recent years, several studies have provided insights into our understanding of C. auris. When C. auris was first isolated from a woman’s ear in 2009, the field was eager to classify it—five diverse clades were identified around the world, but confusion still remained on how it had evolved to be so different from other Candida [8]. It is now understood that C. auris likely evolved in salt marshes, which explains its unique tolerance to high temperatures and high salinity [9]. C. auris infections have also been observed in birds and marine mammals, which suggest possible precursors to human infection [9, 10•]. Additionally, the field’s growing understanding of the skin microbiome—and C. auris’s role within it—has sparked a new understanding of how C. auris colonizes the skin surface, infiltrates the skin through wounds associated with invasive medical devices, and successfully spreads within clinical settings [11, 12••, 13].
Although C. auris demonstrates notable differences as compared with other species in the Candida genus, its phenotypic variation demonstrates a common theme shared with other Candida species and non-Candida pathogenic fungi [14, 15]. C. auris utilizes biofilms, filamentation, and aggregation to adapt to various niches within the body and avoid host immune surveillance [15, 16••]. This review summarizes the incredible phenotypic heterogeneity demonstrated by C. auris and highlights the variants that should be prioritized to advance therapies for this multi-drug-resistant pathogen, which continues to pose a substantial global health risk.
Candida auris Biofilms Colonize Human Skin
Unlike C. albicans and most other Candida species, which are mainly commensal microbes in the gut, vagina, and other internal niches, C. auris cannot survive and persist in an anaerobic environment [17]. However, on the skin, it forms high-burden, multilayer biofilms composed primarily of yeast cells bound by an extracellular matrix [18]. C. auris skin colonization is asymptomatic in over 90% of individuals [19]; still, skin colonization by C. auris is abnormal and represents the greatest single risk factor for the development of candidemia [20]. C. auris is not the only Candida species known to persist in the skin; this group also includes C. tropicalis, C. parapsilosis, and C. orthopsilosis. However, C. auris is relatively more abundant than other Candida species in the skin when present [12••].
Usually, C. auris occupies a commensal role within the skin microbiome; it takes up space so that other more harmful microbes are outcompeted, and therefore, it does not cause disease or clinically significant inflammation. Through direct cell-to-cell interaction and indirect immune modulation, it helps control pathogenic populations and promote diversity and stability in the ecosystem [21].
There is some evidence that the presence of C. auris colonization alters gene expression in the host. In a 3D skin epithelial model, formation of C. auris biofilms was found to induce modest increases in gene expression for several inflammatory cytokines [16••]. In a murine model of skin colonization, C. auris skin colonization triggered a protective IL-17A+ and IL-17F+ response in both innate and adaptive immune cells [22•]. Mice lacking an IL-17 response had a much higher fungal burden, demonstrating that these cytokines play an important role in host control of C. auris.
In addition to host immune modulation in the presence of C. auris colonization, the skin microbiome plays a crucial and largely understudied role in the formation and maintenance of C. auris biofilms in the skin. In a 2021 investigation into the skin microbiomes of patients at a skilled nursing facility, C. auris colonization was associated with a highly dysbiotic microbial profile characterized by the presence of hospital-associated microbes, including Acinetobacter baumannii, Klebsiella pneumoniae, and Enterococcus faecalis. The absence of C. auris colonization was associated with lower levels of Candida and instead the dominance of fungal commensal Malassezia [11]. These findings suggest that C. auris colonization is likely related to skin dysbiosis, which may be induced by antibiotics, antifungals, and disinfectants. Indeed, treatment with antibiotics is a risk factor for C. auris colonization [12••].
C. auris is an efficient colonizer of the skin surface and hair follicles, but it does not typically compromise the skin or invade other tissues spontaneously [18]. Ultimately, C. auris is understood to enter the bloodstream and cause systemic infection mostly through wounds, especially those made by catheters, feeding tubes, and other invasive healthcare equipment [16••]. The artificial wound model used in one study demonstrated that tissues colonized by C. auris exhibited increased cytotoxicity and altered gene expression in response to the induction of a wound [16••]. Of great importance, C. auris is capable of forming persistent and multi-drug-resistant biofilms on surfaces and equipment found in healthcare settings, such as IV poles, axillary probes, lifts/scales, and window curtains [23]. This is a likely mechanism of spread throughout ICUs and nursing homes [4].
In the host skin and on medical equipment, biofilm formation allows for increased resistance to antifungals and desiccation [18, 24]. The increased resistance to antifungals can partially be attributed to the biofilm’s extracellular matrix, which includes a conserved mannan-glucan complex that absorbs antifungals and prevents them from reaching the cells [24]. Another possible mechanism for increased resistance to antifungals in biofilms as compared to planktonic cells is the upregulation of genes encoding efflux pumps, including major facilitator superfamily transporters and ATP-binding cassettes, which may serve to pump antifungals out of vulnerable cells [25].
While understanding C. auris biofilms can provide insights into the pathogenesis and transmission of C. auris infection, its biofilms are typically indicative of a commensal lifestyle. To investigate C. auris–associated candidemia, one must turn to other changes in cellular morphology, including shape and adhesion.
The Filamentation Phenotype of C. auris is Understudied In Vivo
C. auris utilizes distinct morphotypes in different niches within the host to survive, persist, and evade the host immune system [16••]. This strategy is characteristic of the kingdom fungi. One of the most well-conserved phenotypes across fungal genera is the formation of filaments in the shape of hyphae (long, straight lines of cells with continuous, parallel edges) or pseudohyphae (formed by simple budding in one direction) [15]. These long-reaching arms of clonal cells can help invade tightly packed tissues while also being too extensive to be affected by phagocytic cells, such as macrophages and neutrophils. While shared phenotypes contribute to generally comparable invasion and evasion tactics across fungi, there is still significant morphological and functional diversity.
Within the Candida genus, species utilize numerous specialized phenotypes [26, 27]. The most prevalent of these, C. albicans, is found in the upper respiratory, gastrointestinal, and genital tracts, in addition to the skin. C. albicans utilizes a white-opaque bi-stable phenotypic switching system. Each phenotype has a unique metabolic, morphological, and antigenic profile. Opaque cells are more elongated and specialized for skin infections. They perform oxidative metabolism as there is no glucose available on the skin surface. White cells are specialized for systemic and blood infections, performing fermentation in their comparably glucose-rich environment. While both phenotypes are capable of forming pseudohyphae or true hyphae in order to escape phagocytosis or invade new tissues, this response is induced differently in each group [28].
While the white-opaque transition is unique to C. albicans, the ability to form hyphae or pseudohyphae is semi-conserved throughout the genus; C. tropicalis and C. parapsilosis readily form pseudohyphae, while other Candida species form these phenotypic variations more rarely. Notably, C. glabrata rarely forms pseudohyphae and is often found in aggregates or biofilms within the host which allow increased resistance to stress, including antifungals [15]. Across the Candida genus, the formation of hyphae and pseudohyphae is generally associated with more invasive and more virulent behavior [15, 28].
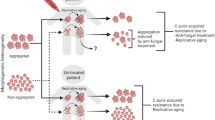
Until a study in 2018, C. auris was widely thought to be filamentation-incompetent, exhibiting only the yeast phenotype (Fig. 1A). In contrast to its relatives in the Candida genus, the filamentation phenotype has not been successfully induced in vitro using any established methods, including exposure to high temperatures, serum, or high CO2 levels [29]. In keeping with the unique halo tolerance of C. auris, however, the formation of structures resembling pseudohyphae had been induced by treatment with 10% NaCl [30]. This encouraged the search for the unique conditions, which might unlock the key to inducing filamentation in C. auris.
In the aforementioned study [29], the morphological transition to a filamentous phenotype could be induced by passage through a mammalian body. After extraction and incubation in vitro, cells recovered from the kidney and liver (but not the brain, spleen, or lungs) demonstrated a 100-fold increase in switching frequency to the filamentous phenotype (Fig. 1B) as compared with the control cells, which had not been passaged through mammalian cells. This capacity for filamentation is heritable; while some cells remained in typical yeast form regardless of condition, cells that switched to the filamentous phenotype were observed to switch between the unicellular yeast and the filamentous phenotype. This switch was bi-directional and was observed in fungal burdens depending on incubation conditions [29]. More recently, another study found that a phenotype resembling filamentous growth can also be induced by genotoxic stress [31]. In both cases, C. auris filaments were found to have both common and distinct characteristics as compared with the hyphae and pseudohyphae found in C. albicans. Specifically, C. auris filaments were found to be multicellular and connected by septa as in C. albicans but are proportionally wider, lack parallel sides, and contain relatively more vacuoles [29, 32•].
The filamentation-competent (FC) yeast [29] also had a notable response to temperature. While in C. albicans, higher temperatures have been observed to induce filamentation, the opposite is observed in C. auris. FC cells incubated at 37 °C were found to revert to typical yeast form, while FC cells incubated at 20 °C maintained their filamentous form in high numbers. This unique response to temperature makes sense in the context of C. auris’s role as a commensal microbe on mammalian skin, which typically maintains a much lower temperature than the internal niche of C. albicans [15, 29]. However, the inhibitory effect of high temperatures on filamentation is common outside the Candida genus, as evidenced by parallel observations in another pathogenic fungus, Histoplasma capsulatum [15].
Gene expression profiles of both unicellular yeast and filamentous phenotypes revealed differential gene expression in numerous metabolic, structural, and regulatory pathways. While filamentous cells exhibit higher rates of transcription for fatty acid metabolites, typical yeast cells demonstrate upregulation in many genes related to glycolysis and the Krebs cycle [29]. The metabolic specialization observed in C. auris resembles that of the white-opaque phenotypic transition in C. albicans. Secreted aspartic proteinases (SAPs), which play a major role in virulence in pathogenic yeasts, were mostly found to be transcribed at similar levels in both yeast and filamentous cells. However, a proteolytic assay found differential SAP activity in each phenotype—filamentous and FC cells demonstrated more SAP activity at higher temperatures, while typical yeast cells demonstrated an inverse relationship [29]. This suggests that unicellular yeast cells in the bloodstream may benefit from post-translational mechanisms, which prevent SAPs from triggering an immune response.
Many filamentation-specific genes with homologs in C. albicans are differentially expressed in a similar pattern to C. auris, with at least two exceptions: EFG1 and CPH2. These genes regulate the white-to-opaque switch in C. albicans but are both downregulated in C. auris [29]. Notably, there are several genes of crucial importance to filamentation in both C. albicans and S. cerevisiae that are completely absent from the genome of C. auris. These include ECE1, HWP1, FLO11, EED1, and HWP2 [32•]. This may partially explain the unique characteristics of C. auris filaments. Interestingly, Tup1, which serves as a transcriptional repressor for filamentous growth in C. albicans, was identified in the genome of C. auris. However, a C. auris Tup1 knockdown mutant did not exhibit increased filamentation, indicating that this pathway is not primarily responsible for the filamentation behavior of C. auris [32•].
While filamentation has been induced in C. auris in vitro, the presence of filamentous cells in vivo in mammalian cells has not been confirmed. It does seem likely, given the findings described above, that epigenetic changes, which promote the filamentous phenotype, are induced upon infection of the liver and kidneys [29]. Since the filamentous phenotype is associated with protection from immune killing in other pathogenic fungi [15], it would make sense for C. auris filaments in the liver and kidneys to serve as defensive reservoirs of C. auris. However, given the inhibitory effect of basal body temperatures on filamentation, it is also possible that C. auris does not convert to the filamentous phenotype within the mammalian host at a high frequency. This is an important topic for future research.
Utilizing the methods to induce filamentation [29], another study found that filamentous strains were more virulent than those of the typical yeast phenotype in a Galleria mellonella infection model [32•]. This makes sense given that filamentation is associated with increased virulence and pathogenicity in most pathogenic fungi, including Candida. However, this finding is difficult to interpret further as the use of a non-mammalian host introduces numerous confounding factors, and the presence of the filamentous phenotype in mammalian cells has not been fully experimentally verified.
The Aggregate Phenotype is Highly Resistant to Antifungals and Attack by the Immune System
The aggregate morphology (Agg) is characterized by large numbers of cells growing in contact with one another (Fig. 1C). This kind of growth can occur within a biofilm or planktonically [33]. Aggregation has been described in several Candida and non-Candida species [33,34,35] and has been implicated in decreased susceptibility to antifungals and increased virulence in the host [34, 36,37,38]. While the aggregate phenotype is considered a subset of filamentous growth in other yeast [33], unicellular yeast cells often form aggregates in C. auris [34]. Aggregation in C. auris demonstrates substantial heterogeneity across its clades and strains, with some strains incapable of forming aggregates (non-Agg) [16••].
In C. auris, the Agg phenotype was first described and noted for its seeming immunity to disruption methods [34]. The pathogenicity and virulence of several common Candida species were assessed in a G. mellonella infection model, finding that C. albicans and C. tropicalis were significantly more virulent than C. parapsilosis and other tested species. Yeast incapable of forming competent pseudohyphae, such as C. glabrata and S. cerevisiae, showed little to no killing of G. mellonella larvae. The exception to this rule was C. auris—although dissections of infected larvae revealed no pseudohyphal growth, the non-Agg phenotype was as virulent as C. albicans, the most virulent of assessed Candida species. The Agg phenotype was observed to preserve its aggregates in vivo but was significantly less virulent than its non-Agg counterpart [34].
A 2020 study sought to further compare pathogenicity and gene expression of C. auris in the Agg and non-Agg phenotypes using in vitro wound models. More genes were found to be upregulated in the Agg than the non-Agg phenotype, across both planktonic and biofilm states. In addition to the difference in quantity, there was also a significant difference in quality—in Agg biofilms, most upregulated genes were structural, including TSA1, ECM33, MP65, PHR1, and several adhesins including ALS1. These genes have been found to play important roles in aggregation and biofilm formation in C. albicans. These changes make sense as possible contributors to cellular “stickiness.” Conversely, non-Agg biofilms exhibited the most increase in expression for genes related to metabolism and biological processes [16••].
Of clinical importance, Agg and non-Agg C. auris showed similar low cytotoxicity on an intact skin model, and only after the induction of the wound did these phenotypes become highly cytotoxic and induced inflammation in the tissue. Following the induction of the wound, the Agg phenotype was significantly more cytotoxic and pro-inflammatory as compared with the non-Agg phenotype [16••].
This study paints a contradictory picture compared to the first study that described the Agg phenotype [34]. The non-Agg phenotype of C. auris is more virulent when inoculated, but Agg C. auris has more potential for invasive growth and immunogenicity when introduced via a skin wound. Both have the potential to participate in biofilms and persist commensally on the skin. These observations are consistent with the concept of phenotypic plasticity in yeast. The human host is composed of numerous distinct niches, and to mount a competent infection across these changing conditions, C. auris benefits from shifting between different morphotypes and antigenic profiles. Unicellular, non-Agg, planktonic C. auris is in stealth mode; it is virtually undetectable by innate immune cells [39] and has a rather unique transcriptional profile that shares minimal similarity to other pathogenic yeast [16••]. Conversely, the Agg phenotype of C. auris represents a more heavy-duty, defensive phenotype. For the cost of increased immunogenicity [16••], it gains increased resistance to members of all known classes of antifungals with significant heterogeneity across clades and strains [35]. This is supported by the fact that even low-level exposures to triazole and echinocandin antifungals strongly and reversibly induce aggregate formation [35].
Stress may also play a role in the induction of the Agg phenotype. In all known fungal pathogens, Hog1, a conserved stress-activated protein kinase, has been shown to be crucial to virulence in vivo [17]. Interestingly, C. auris Hog1 knockdown mutants were found to form large aggregates, which exhibited increased resistance to antimicrobials targeting the cell wall, decreased virulence in a C. elegans model, and decreased tolerance to osmotic, oxidative, and SDS-imposed stresses. While Hog1 may play a role in aggregate formation, these knockdown mutants are hardly a replicate of the aggregate phenotype, as they exhibit decreased stress tolerance and are easily disturbed, in contrast to the physically tough and highly stress-tolerant wild-type Agg phenotype. Still, Hog1 plays a crucial role in C. auris virulence as it does in other fungal pathogens and may relate to the decreased virulence of the Agg phenotype [17].
A recent investigation into C. auris aggregation distinguished typical aggregation from antifungal-induced aggregation [40]. In Agg-competent strains, aggregation can be induced by nutrient-rich media and is primarily mediated by the Als-family adhesin found in C. albicans. In contrast, aggregation induced by antifungals was found to be caused by defects in cell separation—this is referred to as clustering. This finding has major implications for differences in gene expression between the two visually similar phenotypes. However, both the true Agg phenotype and the clustering phenotype share functional similarity in their increased resistance to antifungals and their immunity to clearing by macrophages [40].
The aggregate phenotype provides an opportunity for C. auris to survive and persist within the body even in the face of immune surveillance and treatment with antifungals. Aggregates of C. auris have been identified within the kidney tissue of mice killed by candidemia [41], further suggesting that aggregates may serve as a persistent reservoir of infection. As such, the aggregate phenotype represents a crucial target for further study and treatment.
Conclusions
The ability to undergo microevolution in the host is important to the survival and persistence of microbial pathogens. For many members of the human microbiome, changes in phenotype can influence the shift between commensal and pathogenic lifestyles. Phenotypic variation is caused by changes in gene expression, which might correspond to altered metabolic pathways, modified susceptibility to drugs, and differential antigenicity.
C. auris has acquired several unique phenotypes in the relatively short amount of time it has interacted with and adapted to its host. The identification of five geographically and phylogenetically distinct clades of C. auris [8] represents only one facet of its diversity. While different strains may demonstrate varying abilities to form filaments and aggregates [34], a single strain may also exhibit significant morphological plasticity in response to various conditions [29]. Given this functional and morphological diversity, unraveling the phenotypic switches employed by C. auris in vitro and in vivo remains difficult.
Based on what has been observed in C. auris and other pathogenic fungi within and outside of the Candida genus, we can make some predictions about the functional role of each unique phenotype. It is understood that C. auris forms robust and high-burden biofilms on the skin [18]. The presence of C. auris is associated with an altered microbial profile [11]. Conditions found on the human skin, including lower temperatures and the presence of salt in the form of sweat, promote filamentation in some C. auris strains [29]. Filamentation is associated with increased invasiveness, especially in hair follicles, which may serve as a reservoir of C. auris colonization, allowing it to persist on the skin for many months undetected [22•]. Regardless of the presence of biofilms or filamentation, C. auris likely enters the bloodstream mostly through wounds, including those created by invasive medical equipment [16••]. High temperatures in the blood are associated with the yeast phenotype, which is the most prevalent, virulent, and least immunogenic form of C. auris, promoting systemic infection [29]. The aggregate phenotype is perhaps the most variable between strains [35] but can be triggered by environmental conditions or exposure to antifungals [40]. Having been found in the kidneys of C. auris–killed mice, aggregates may also serve as a reservoir for C. auris infection [41].
Of great importance in future studies is the utilization of mammalian hosts, as temperature and immunogenicity have a large influence on fungal phenotypic switching and the transition from commensal to pathogenic lifestyles. More in vivo experiments might prove increasingly crucial to an understanding of the morphological plasticity and dynamic nature of C. auris infection.
More research should be done exploring C. auris as a commensal species and considering how the interaction of C. auris with other commensal skin microbes, including bacteria and Malassezia, might influence the transition to pathogenicity. Since the presence of Malassezia on the skin is associated with decreased Candida, including C. auris [12••], it may prove an important target for promoting a resilient skin microbiome. The role of sweat in inducing the filamentous phenotype and therefore increasing virulence should also be a focus of future experiments, as this may serve as a viable target for avoiding transmission from the skin surface to the bloodstream.
Given the robust resistance to antifungals found in many strains of C. auris, the creation of new antifungal drugs may improve the scope of treatment options for candidemia. However, this is a slippery slope, as the use of antifungals continues to select for more drug-resistant pathogenic fungi, which already exist in the environment [42]. Additionally, the effects of antifungal drugs on C. auris may result in increased antifungal and immune resistance, as in the case of the aggregate phenotype triggered by antifungal exposure [40].
As an alternative to new antifungals, reduction of invasive procedures/devices and the exploration of noninvasive alternatives should be considered, as wound induction is a major factor in C. auris pathogenesis. Updating hygiene practices and disinfection of medical equipment might also reduce the likelihood of C. auris spread among ICUs and nursing homes.
C. auris represents a unique opportunity to study the rise of a novel pathogen. Given the fact that climate change is causing rising temperatures in many regions across the globe and that this in turn selects for more thermotolerant fungi, it is likely that more novel fungal and non-fungal pathogens will arrive on the scene in the years to come. Because C. auris is a thermo- and halo-tolerant fungus, which has only recently evolved to infect the mammalian host, and because it demonstrates remarkable resistance to antifungals, it is truly a fungus for our time, emblematic of the issues that face the medical field as it progresses further into the twenty-first century.
Data Availability
Not applicable.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Centers for Disease Control and Prevention. 2022 [cited 2023 Jul 24]. The biggest antibiotic-resistant threats in the U.S. Available from: https://www.cdc.gov/drugresistance/biggest-threats.html. Accessed 8/18/2023
Ricotta EE, Lai YL, Babiker A, Strich JR, Kadri SS, Lionakis MS, et al. Invasive candidiasis species distribution and trends, United States, 2009–2017. J Infect Dis. 2021;223(7):1295–302.
Alvarez-Moreno CA, Morales-López S, Rodriguez GJ, Rodriguez JY, Robert E, Picot C, et al. The mortality attributable to candidemia in C. auris is higher than that in other Candida species: Myth or Reality? J Fungi Basel Switz. 2023;9(4):430.
Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018;379(14):1322–31.
Oremefetse D, Aijaz A, Sanelisiwe D, Mrudula P. Survival of Candida auris on environmental surface material and low-level resistance to disinfectant. J Hosp Infect. 2023;S0195-6701(23):00120–2.
Bagheri Lankarani K, Akbari M, Tabrizi R, Vali M, Sekhavati E, Heydari ST, et al. Candida auris: outbreak fungal pathogen in COVID-19 pandemic: a systematic review and meta-analysis. Iran J Microbiol. 2022;14(3):276–84.
Casadevall A. Climate change brings the specter of new infectious diseases. J Clin Invest. 2020;130(2):553–5.
Du H, Bing J, Hu T, Ennis CL, Nobile CJ, Huang G. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16(10):e1008921.
Arora P, Singh P, Wang Y, Yadav A, Pawar K, Singh A, et al. Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India. mBio. 2021; https://doi.org/10.1128/mBio.03181-20.
Garcia-Bustos V, Cabañero-Navalon MD, Ruiz-Gaitán A, Salavert M, Tormo-Mas MÁ, Pemán J. Climate change, animals, and Candida auris: insights into the ecological niche of a new species from a One Health approach. Clin Microbiol Infect. https://www.sciencedirect.com/science/article/pii/S1198743X23001325. Accessed 8/18/2023. This paper summarizes recent findings related to the ecological niche of Candida auris in wetland environments, which has major implications for the origins of C. auris’s unique phenotypic heterogeneity, halo- and thermotolerance, and its antifungal resistance.
Huang X, Welsh RM, Deming C, Proctor DM, Thomas PJ, Comparative Sequencing Program NISC, et al. Skin metagenomic sequence analysis of early Candida auris outbreaks in U.S. nursing homes. mSphere. 2021;6(4):e0028721.
Proctor DM, Dangana T, Sexton DJ, Fukuda C, Yelin RD, Stanley M, et al. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med. 2021;27(8):1401–9. This paper provides crucial insights into the microbial ecology of Candida auris infection on the skin surface.
Tharp B, Zheng R, Bryak G, Litvintseva AP, Hayden MK, Chowdhary A, et al. Role of microbiota in the skin colonization of Candida auris. mSphere. 8(1):e00623.
Bouklas T, Jain N, Fries BC. Modulation of replicative lifespan in Cryptococcus neoformans: implications for virulence. Front Microbiol. 2017;30(8):98.
Min K, Neiman AM, Konopka JB. Fungal pathogens: shape-shifting invaders. Trends Microbiol. 2020;28(11):922–33.
Brown JL, Delaney C, Short B, Butcher MC, McKloud E, Williams C, et al. Candida auris phenotypic heterogeneity determines pathogenicity in vitro. mSphere. 2020;5(3):e00371. This paper characterizes important differences in pathogenicity and gene expression across aggregative and non-aggregative strains of C. auris. It also includes crucial evidence for the role of wounds in C. auris candidemia pathogenesis.
Day AM, McNiff MM, da Silva DA, Gow NAR, Quinn J. Hog1 Regulates stress tolerance and virulence in the emerging fungal pathogen Candida auris. mSphere. 2018;3(5) https://doi.org/10.1128/msphere.00506-18.
Horton MV, Johnson CJ, Kernien JF, Patel TD, Lam BC, Cheong JZA, et al. Candida auris forms high-burden biofilms in skin niche conditions and on porcine skin. mSphere. 2020;5(1):e00910–9.
Rubin R. On the Rise, Candida auris outwits treatments and travels incognito in health care settings. JAMA. 2023;329(3):197–9.
Vallabhaneni S. Investigation of the First seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus — United States, May 2013–August 2016. MMWR Morb Mortal Wkly Rep. 2016;65 https://www.cdc.gov/mmwr/volumes/65/wr/mm6544e1.htm. Accessed 8/18/2023
Proctor DM, Drummond RA, Lionakis MS, Segre JA. One population, multiple lifestyles: commensalism and pathogenesis in the human mycobiome. Cell Host Microbe. 2023;31(4):539–53.
Huang X, Hurabielle C, Drummond RA, Bouladoux N, Desai JV, Sim CK, et al. Murine model of colonization with fungal pathogen Candida auris to explore skin tropism, host risk factors and therapeutic strategies. Cell Host Microbe. 2021;29(2):210–221.e6. This in vivo mouse model of skin infection further characterizes C. auris biofilms on mammalian skin, revealing their extreme longevity and resilience.
Adams E, Quinn M, Tsay S, Poirot E, Chaturvedi S, Southwick K, et al. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg Infect Dis. 2018;24(10):1816–24.
Dominguez EG, Zarnowski R, Choy HL, Zhao M, Sanchez H, Nett JE, et al. Conserved Role for biofilm matrix polysaccharides in Candida auris drug resistance. mSphere. 2019;4(1):e00680–18.
Kean R, Delaney C, Sherry L, Borman A, Johnson EM, Richardson MD, et al. Transcriptome assembly and profiling of Candida auris reveals novel insights into biofilm-mediated resistance. mSphere. 2018;3(4):e00334.
Polke M, Hube B, Jacobsen ID. Candida survival strategies. Adv Appl Microbiol. 2015;91:139–235.
Bouklas T, Alonso-Crisóstomo L, Székely T, Diago-Navarro E, Orner EP, Smith K, et al. Generational distribution of a Candida glabrata population: resilient old cells prevail, while younger cells dominate in the vulnerable host. PLoS Pathog. 2017;13(5):e1006355.
Bommanavar SB, Gugwad S, Malik N. Phenotypic switch: the enigmatic white-gray-opaque transition system of Candida albicans. J Oral Maxillofac Pathol JOMFP. 2017;21(1):82–6.
Yue H, Bing J, Zheng Q, Zhang Y, Hu T, Du H, et al. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect. 2018;7:188.
Wang X, Bing J, Zheng Q, Zhang F, Liu J, Yue H, et al. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect. 2018;7(1):93.
Ruiz G, Ross Z, Gow N, Lorenz A. Pseudohyphal growth of the emerging pathogen Candida auris is triggered by genotoxic stress through the S phase checkpoint. mSphere. 2020;5:e00151-20
Fan S, Yue H, Zheng Q, Bing J, Tian S, Chen J, et al. Filamentous growth is a general feature of Candida auris clinical isolates. Med Mycol. 2021;59(7):734–40. This paper characterizes filamentous growth in terms of its antifungal resistance and virulence in a Galleria mellonella infection model.
Chow J, Dionne HM, Prabhakar A, Mehrotra A, Somboonthum J, Gonzalez B, et al. Aggregate filamentous growth responses in yeast. mSphere. 2019;4(2):e00702–18.
Borman AM, Szekely A, Johnson EM. Comparative pathogenicity of United Kingdom Isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. mSphere. 2016;1(4):e00189.
Szekely A, Borman AM, Johnson EM. Candida auris isolates of the Southern Asian and South African lineages exhibit different phenotypic and antifungal susceptibility profiles in vitro. J Clin Microbiol. 2019;57(5):e02055.
Sherry L, Ramage G, Kean R, Borman A, Johnson EM, Richardson MD, et al. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis. 2017;23(2):328–31.
Short B, Brown J, Delaney C, Sherry L, Williams C, Ramage G, et al. Candida auris exhibits resilient biofilm characteristics in vitro: implications for environmental persistence. J Hosp Infect. 2019;103(1):92–6.
Singh R, Kaur M, Chakrabarti A, Shankarnarayan SA, Rudramurthy SM. Biofilm formation by Candida auris isolated from colonising sites and candidemia cases. Mycoses. 2019;62(8):706–9.
Johnson CJ, Davis JM, Huttenlocher A, Kernien JF, Nett JE. Emerging fungal pathogen Candida auris evades neutrophil attack. mBio. 2018;9(4):e01403–18.
Pelletier C, Brown AJP, Lorenz A. Candida auris undergoes adhesin-dependent and -independent cellular aggregation. bioRxiv. 2023; https://doi.org/10.1101/2023.04.21.537817v1.
Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, et al. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 2017;23(2):195–203.
Varma A, Kwon-Chung KJ. Heteroresistance of Cryptococcus gattii to Fluconazole. Antimicrob Agents Chemother. 2010;54(6):2303–11.
Author information
Authors and Affiliations
Contributions
J.C.S. and T.B. wrote the manuscript. D.R.G. created the figure and wrote some sections of the manuscript. T.B. revised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Competing Interests
The authors declare no competing interest.
Human/Animal Studies Informed Consent Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of Topical Collection on Tropical Mycoses
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stephenson, J.C., Garza, D.R. & Bouklas, T. A Fungus for Our Time: Candida auris Emerges into the Anthropocene. Curr Trop Med Rep 10, 244–251 (2023). https://doi.org/10.1007/s40475-023-00293-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40475-023-00293-w