Abstract
Purpose of Review
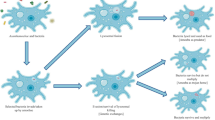
Free-living amoebae (FLAs) are ubiquitous and can co-habit similar niches and interact with fungi. Herein, we discuss theories on FLAs and the origin, evolution, and conservation of fungal virulence, proposing the “feast-fit-fist-feat” hypothesis that covers the knowledge on FLA-fungi interactions, and could be extended during evolutionarily host escalation. Overall, by bridling this selective pressure, fungi might return to environment and by serendipity, infect superior hosts. The selected traits might grant the fungus with an enhanced capacity to cause damage, or virulence. The fungal virulence factors that might be expressed during infection to amoeba and that grant a fungal benefit during infection to mammals are discussed. However, how they are induced during infection of FLAs is still an open field. Here we discuss also the “Trojan Horse” role of FLAs and the importance of co-infections and disease outcome.
Recent Findings
Herein, we discuss also at the molecular level the early steps on how FLAs are able to attach and internalize fungal pathogens. Upon entrance, amoeba interaction might pose selective pressures, and the result is usually a more virulent phenotype of the fungus. Amoeba is able to modulate several fungal virulence factors, most of them with relative importance for infection to superior or more evolved hosts. This interaction fungi-FLAs makes an attractive model for the application of the “One Health” concept in order to avoid new emerging more virulent fungal species.
Summary
Amoeba-fungi interactions are still an open field, with several avenues yet to be explored, which might explain the origin of microbial virulence and innate immunity evolution. Several mechanisms of direct or indirect regulation might be involved.




Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: •• Of major importance
Armand B, Motazedian MH, Asgari Q. Isolation and identification of pathogenic free-living amoeba from surface and tap water of Shiraz City using morphological and molecular methods. Parasitol Res. 2016;115(1):63–8. https://doi.org/10.1007/s00436-015-4721-7.
Denet E, Coupat-Goutaland B, Nazaret S, Pelandakis M, Favre-Bonte S. Diversity of free-living amoebae in soils and their associated human opportunistic bacteria. Parasitol Res. 2017;116(11):3151–62. https://doi.org/10.1007/s00436-017-5632-6.
Gomes Tdos S, Magnet A, Izquierdo F, Vaccaro L, Redondo F, Bueno S, et al. Acanthamoeba spp. in contact lenses from healthy individuals from Madrid, Spain. PLoS One. 2016;11(4):e0154246. https://doi.org/10.1371/journal.pone.0154246.
Reyes-Batlle M, Wagner C, Zamora-Herrera J, Vargas-Mesa A, Sifaoui I, Gonzalez AC, et al. Isolation and molecular identification of Vermamoeba vermiformis strains from soil sources in El Hierro Island, Canary Islands, Spain. Curr Microbiol. 2016;73(1):104–7. https://doi.org/10.1007/s00284-016-1035-7.
Soares SS, Souza TK, Berte FK, Cantarelli VV, Rott MB. Occurrence of infected free-living amoebae in cooling towers of southern Brazil. Curr Microbiol. 2017;74(12):1461–8. https://doi.org/10.1007/s00284-017-1341-8.
Ustunturk-Onan M, Walochnik J. Identification of free-living amoebae isolated from tap water in Istanbul, Turkey. Exp Parasitol. 2018;195:34–7. https://doi.org/10.1016/j.exppara.2018.10.002.
Wanner M, Birkhofer K, Puppe D, Shimano SD, Shimizu M. Tolerance of testate amoeba species to rising sea levels under laboratory conditions and in the South Pacific. Pedobiologia. 2020;79:150610. https://doi.org/10.1016/j.pedobi.2019.150610.
Wopereis DB, Bazzo ML, de Macedo JP, Casara F, Golfeto L, Venancio E, et al. Free-living amoebae and their relationship to air quality in hospital environments: characterization of Acanthamoeba spp. obtained from air-conditioning systems. Parasitology. 2020;147(7):782–90. https://doi.org/10.1017/S0031182020000487.
Guimaraes AJ, Gomes KX, Cortines JR, Peralta JM, Peralta RH. Acanthamoeba spp. as a universal host for pathogenic microorganisms: one bridge from environment to host virulence. Microbiol Res. 2016;193:30–8. https://doi.org/10.1016/j.micres.2016.08.001.
Magnet A, Galvan AL, Fenoy S, Izquierdo F, Rueda C, Fernandez Vadillo C, et al. Molecular characterization of Acanthamoeba isolated in water treatment plants and comparison with clinical isolates. Parasitol Res. 2012;111(1):383–92. https://doi.org/10.1007/s00436-012-2849-2.
Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17(2):413–33. https://doi.org/10.1128/cmr.17.2.413-433.2004.
Casadevall A, Fu MS, Guimaraes AJ, Albuquerque P. The ‘Amoeboid Predator-Fungal Animal Virulence’ hypothesis. J Fungi (Basel). 2019;5(1). https://doi.org/10.3390/jof5010010.
Hillmann F, Novohradska S, Mattern DJ, Forberger T, Heinekamp T, Westermann M, et al. Virulence determinants of the human pathogenic fungus Aspergillus fumigatus protect against soil amoeba predation. Environ Microbiol. 2015;17(8):2858–69. https://doi.org/10.1111/1462-2920.12808.
Khan NA. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol Rev. 2006;30(4):564–95. https://doi.org/10.1111/j.1574-6976.2006.00023.x.
Goncalves DS, MDS F, Gomes KX, Rodriguez-de La Noval C, Liedke SC, da Costa GCV, et al. Unravelling the interactions of the environmental host Acanthamoeba castellanii with fungi through the recognition by mannose-binding proteins. Cell Microbiol. 2019;21(10):e13066. https://doi.org/10.1111/cmi.13066. Description of fungi-amoeba interactions at the molecular level and the participation of mannose-binding receptors.
Samba-Louaka A, Delafont V, Rodier MH, Cateau E, Hechard Y. Free-living amoebae and squatters in the wild: ecological and molecular features. FEMS Microbiol Rev. 2019;43(4):415–34. https://doi.org/10.1093/femsre/fuz011.
Krol-Turminska K, Olender A. Human infections caused by free-living amoebae. Ann Agric Environ Med. 2017;24(2):254–60. https://doi.org/10.5604/12321966.1233568.
Casadevall A. Evolution of intracellular pathogens. Annu Rev Microbiol. 2008;62:19–33. https://doi.org/10.1146/annurev.micro.61.080706.093305.
Escoll P, Rolando M, Gomez-Valero L, Buchrieser C. From amoeba to macrophages: exploring the molecular mechanisms of Legionella pneumophila infection in both hosts. Curr Top Microbiol Immunol. 2013;376:1–34. https://doi.org/10.1007/82_2013_351.
Darwin C. On The Origin of Species by Means of Natural Selection, or Preservation of Favoured Races in the Struggle for Life. London: John Murray. 1809-1882; 1859.
Berger J. Ways of seeing. New York: Viking Press; 1973.
Casadevall A. Amoeba provide insight into the origin of virulence in pathogenic fungi. Adv Exp Med Biol. 2012;710:1–10. https://doi.org/10.1007/978-1-4419-5638-5_1.
Novohradska S, Ferling I, Hillmann F. Exploring virulence determinants of filamentous fungal pathogens through interactions with soil amoebae. Front Cell Infect Microbiol. 2017;7:497. https://doi.org/10.3389/fcimb.2017.00497. One of the most complete reviews explaining the evolutionary emergence and selection of the virulence factors of filamentous in soil.
Bertelli C, Greub G. Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front Cell Infect Microbiol. 2012;2:110. https://doi.org/10.3389/fcimb.2012.00110.
Casadevall A, Steenbergen JN, Nosanchuk JD. ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi--the Cryptococcus neoformans paradigm. Curr Opin Microbiol. 2003;6(4):332–7. https://doi.org/10.1016/s1369-5274(03)00082-1.
Zaragoza O. Basic principles of the virulence of Cryptococcus. Virulence. 2019;10(1):490–501. https://doi.org/10.1080/21505594.2019.1614383.
Bozzaro S, Eichinger L. The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets. 2011;12(7):942–54. https://doi.org/10.2174/138945011795677782.
Jose Maschio V, Corcao G, Rott MB. Identification of Pseudomonas spp. as amoeba-resistant microorganisms in isolates of Acanthamoeba. Rev Inst Med Trop Sao Paulo. 2015;57(1):81–3. https://doi.org/10.1590/S0036-46652015000100012.
Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165):165rv13. https://doi.org/10.1126/scitranslmed.3004404.
Albuquerque P, Nicola AM, Magnabosco DAG, Derengowski LDS, Crisostomo LS, Xavier LCG, et al. A hidden battle in the dirt: soil amoebae interactions with Paracoccidioides spp. PLoS Negl Trop Dis. 2019;13(10):e0007742. https://doi.org/10.1371/journal.pntd.0007742.
Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba. Parasit Vectors. 2012;5:6. https://doi.org/10.1186/1756-3305-5-6.
Mulec J, Dietersdorfer E, Ustunturk-Onan M, Walochnik J. Acanthamoeba and other free-living amoebae in bat guano, an extreme habitat. Parasitol Res. 2016;115(4):1375–83. https://doi.org/10.1007/s00436-015-4871-7.
Azzam SZ, Cayme GJ, Martinez LR. Polymicrobial interactions involving fungi and their importance for the environment and in human disease. Microb Pathog. 2020;140:103942. https://doi.org/10.1016/j.micpath.2019.103942.
Grice EA, Dawson TL Jr. Host-microbe interactions: Malassezia and human skin. Curr Opin Microbiol. 2017;40:81–7. https://doi.org/10.1016/j.mib.2017.10.024.
Heaselgrave W, Shama G, Andrew PW, Kong MG. Inactivation of Acanthamoeba spp. and other ocular pathogens by application of cold atmospheric gas plasma. Appl Environ Microbiol. 2016;82(10):3143–8. https://doi.org/10.1128/AEM.03863-15.
Segal E, Frenkel M. Dermatophyte infections in environmental contexts. Res Microbiol. 2015;166(7):564–9. https://doi.org/10.1016/j.resmic.2014.12.007.
Buchele MLC, Wopereis DB, Casara F, de Macedo JP, Rott MB, Monteiro FBF, et al. Contact lens-related polymicrobial keratitis: Acanthamoeba spp. genotype T4 and Candida albicans. Parasitol Res. 2018;117(11):3431–6. https://doi.org/10.1007/s00436-018-6037-x.
Kilvington S, Lonnen J. A comparison of regimen methods for the removal and inactivation of bacteria, fungi and Acanthamoeba from two types of silicone hydrogel lenses. Cont Lens Anterior Eye. 2009;32(2):73–7. https://doi.org/10.1016/j.clae.2008.12.008.
Martinez AJ. Acanthamoebiasis and immunosuppression. Case report. J Neuropathol Exp Neurol. 1982;41(5):548–57. https://doi.org/10.1097/00005072-198209000-00007.
Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A. 2001;98(26):15245–50. https://doi.org/10.1073/pnas.261418798.
Castellani A. An amoeba growing in cultures of a yeast. J Trop Med Hyg. 1930;33:188–91.
Volkonsky M. Hartmannella castellanii Douglas et classification des Hartmannelles. Archiv Zool Exp Generale. 1931;72:317–39.
Castellani A. Phagocytic and destructive action of Hartmanella castellanii (Amoeba castellanii) on pathogenic encapsulated yeast-like fungi Torulopsis neoformans (Cryptococcus neoformans). Ann Inst Pasteur (Paris). 1955;89(1):1–7.
Nero LC, Tarver MG, Hedrick LR. Growth of Acanthamoeba castellani with the yeast Torulopsis Famata. J Bacteriol. 1964;87:220–5. https://doi.org/10.1128/JB.87.1.220-225.1964.
Bunting LA, Neilson JB, Bulmer GS. Cryptococcus neoformans: gastronomic delight of a soil ameba. Sabouraudia. 1979;17(3):225–32. https://doi.org/10.1080/00362177985380341.
Ruiz A, Neilson JB, Bulmer GS. Control of Cryptococcus neoformans in nature by biotic factors. Sabouraudia. 1982;20(1):21–9.
Neilson JB, Fromtling RA, Bulmer GS. Pseudohyphal forms of Cryptococcus neoformans: decreased survival in vivo. Mycopathologia. 1981;73(1):57–9. https://doi.org/10.1007/BF00443015.
Chrisman CJ, Alvarez M, Casadevall A. Phagocytosis of Cryptococcus neoformans by, and nonlytic exocytosis from, Acanthamoeba castellanii. Appl Environ Microbiol. 2010;76(18):6056–62. https://doi.org/10.1128/AEM.00812-10.
Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. Interaction of Blastomyces dermatitidis, Sporothrix schenckii, and Histoplasma capsulatum with Acanthamoeba castellanii. Infect Immun. 2004;72(6):3478–88. https://doi.org/10.1128/IAI.72.6.3478-3488.2004.
Radosa S, Ferling I, Sprague JL, Westermann M, Hillmann F. The different morphologies of yeast and filamentous fungi trigger distinct killing and feeding mechanisms in a fungivorous amoeba. Environ Microbiol. 2019;21(5):1809–20. https://doi.org/10.1111/1462-2920.14588. Manuscript showing how distinct fungal morphologies can determine the fungal fate in fungivorous amoeba.
Drummond RA, Lionakis MS. Mechanistic insights into the role of C-type lectin receptor/CARD9 signaling in human antifungal immunity. Front Cell Infect Microbiol. 2016;6:39. https://doi.org/10.3389/fcimb.2016.00039.
Nikolakopoulou C, Willment JA, Brown GD. C-Type Lectin Receptors in Antifungal Immunity. Adv Exp Med Biol. 2020;1204:1-30. https://doi.org/10.1007/978-981-15-1580-41.
Plato A, Hardison SE, Brown GD. Pattern recognition receptors in antifungal immunity. Semin Immunopathol. 2015;37(2):97–106. https://doi.org/10.1007/s00281-014-0462-4.
Brown GD. Innate antifungal immunity: the key role of phagocytes. Annu Rev Immunol. 2011;29:1–21. https://doi.org/10.1146/annurev-immunol-030409-101229.
Esher SK, Zaragoza O, Alspaugh JA. Cryptococcal pathogenic mechanisms: a dangerous trip from the environment to the brain. Mem Inst Oswaldo Cruz. 2018;113(7):e180057. https://doi.org/10.1590/0074-02760180057.
Watkins RA, Andrews A, Wynn C, Barisch C, King JS, Johnston SA. Cryptococcus neoformans escape from dictyostelium amoeba by both WASH-mediated constitutive exocytosis and Vomocytosis. Front Cell Infect Microbiol. 2018;8:108. https://doi.org/10.3389/fcimb.2018.00108.
Bowen ID, Coakley WT, James CJ. The digestion of Saccharomyces cerevisiae by Acanthamoeba castellanii. Protoplasma. 1979;98(1–2):63–71.
Lemos Tavares P, Carvalho Ribeiro A, Kercher Berte F, da Silva Hellwig AH, Machado Pagani D. Tavares de Souza CC et al. the interaction between Sporothrix schenckii sensu stricto and Sporothrix brasiliensis with Acanthamoeba castellanii. Mycoses. 2020;63(3):302–7. https://doi.org/10.1111/myc.13043.
Ferling I, Dunn JD, Ferling A, Soldati T, Hillmann F. Conidial melanin of the human-pathogenic fungus Aspergillus fumigatus disrupts cell autonomous defenses in amoebae. mBio. 2020;11(3). https://doi.org/10.1128/mBio.00862-20. Importance of melanin for fungal protection during interactions with amoeba.
Hubert F, Rodier MH, Minoza A, Portet-Sulla V, Cateau E, Brunet K. Free-living amoebae promote Candida auris survival and proliferation in water. Lett Appl Microbiol. 2020. https://doi.org/10.1111/lam.13395.
Rizzo J, Albuquerque PC, Wolf JM, Nascimento R, Pereira MD, Nosanchuk JD, et al. Analysis of multiple components involved in the interaction between Cryptococcus neoformans and Acanthamoeba castellanii. Fungal Biol. 2017;121(6–7):602–14. https://doi.org/10.1016/j.funbio.2017.04.002.
Nunes TE, Brazil NT, Fuentefria AM, Rott MB. Acanthamoeba and Fusarium interactions: a possible problem in keratitis. Acta Trop. 2016;157:102–7. https://doi.org/10.1016/j.actatropica.2016.02.001.
Cateau E, Hechard Y, Fernandez B, Rodier MH. Free living amoebae could enhance Fusarium oxysporum growth. Fungal Ecol. 2014;8:12–7.
Maisonneuve E, Cateau E, Kaaki S, Rodier MH. Vermamoeba vermiformis-Aspergillus fumigatus relationships and comparison with other phagocytic cells. Parasitol Res. 2016;115(11):4097–105. https://doi.org/10.1007/s00436-016-5182-3.
Fu MS, Casadevall A. Divalent metal cations potentiate the predatory capacity of amoeba for Cryptococcus neoformans. Appl Environ Microbiol. 2018;84(3). https://doi.org/10.1128/AEM.01717-17.
Madu UL, Ogundeji AO, Mochochoko BM, Pohl CH, Albertyn J, Swart CW, et al. Cryptococcal 3-hydroxy fatty acids protect cells against amoebal phagocytosis. Front Microbiol. 2015;6:1351. https://doi.org/10.3389/fmicb.2015.01351.
Madu UL, Ogundeji AO, Pohl CH, Albertyn J, Sebolai OM. Elucidation of the role of 3-hydroxy fatty acids in Cryptococcus-amoeba interactions. Front Microbiol. 2017;8:765. https://doi.org/10.3389/fmicb.2017.00765.
Casadevall A. Determinants of virulence in the pathogenic fungi. Fungal Biol Rev. 2007;21(4):130–2. https://doi.org/10.1016/j.fbr.2007.02.007.
Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6(12):2169–74. https://doi.org/10.1128/EC.00308-07.
Singulani JL, Scorzoni L, de Oliveira HC, Marcos CM, Assato PA, Fusco-Almeida AM, et al. Applications of invertebrate animal models to dimorphic fungal infections. J Fungi (Basel). 2018;4(4). https://doi.org/10.3390/jof4040118.
Koller B, Schramm C, Siebert S, Triebel J, Deland E, Pfefferkorn AM, et al. Dictyostelium discoideum as a novel host system to study the interaction between phagocytes and yeasts. Front Microbiol. 2016;7:1665. https://doi.org/10.3389/fmicb.2016.01665.
Van Waeyenberghe L, Bare J, Pasmans F, Claeys M, Bert W, Haesebrouck F, et al. Interaction of Aspergillus fumigatus conidia with Acanthamoeba castellanii parallels macrophage-fungus interactions. Environ Microbiol Rep. 2013;5(6):819–24. https://doi.org/10.1111/1758-2229.12082. One of the most important manuscripts demonstrating similarities on the mechanisms of interaction of amoeba and macrophage with fungi.
Caza M, Kronstad JW. The cAMP/protein kinase a pathway regulates virulence and adaptation to host conditions in Cryptococcus neoformans. Front Cell Infect Microbiol. 2019;9:212. https://doi.org/10.3389/fcimb.2019.00212.
Hommel B, Mukaremera L, Cordero RJB, Coelho C, Desjardins CA, Sturny-Leclere A, et al. Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog. 2018;14(5):e1006982. https://doi.org/10.1371/journal.ppat.1006982.
Maliehe M, Ntoi MA, Lahiri S, Folorunso OS, Ogundeji AO, Pohl CH, et al. Environmental factors that contribute to the maintenance of Cryptococcus neoformans pathogenesis. Microorganisms. 2020;8(2). https://doi.org/10.3390/microorganisms8020180.
Hu G, Cheng PY, Sham A, Perfect JR, Kronstad JW. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol. 2008;69(6):1456–75. https://doi.org/10.1111/j.1365-2958.2008.06374.x.
Derengowski Lda S, Paes HC, Albuquerque P, Tavares AH, Fernandes L, Silva-Pereira I, et al. The transcriptional response of Cryptococcus neoformans to ingestion by Acanthamoeba castellanii and macrophages provides insights into the evolutionary adaptation to the mammalian host. Eukaryot Cell. 2013;12(5):761–74. https://doi.org/10.1128/EC.00073-13.
Gerstein AC, Jackson KM, McDonald TR, Wang Y, Lueck BD, Bohjanen S, et al. Identification of pathogen genomic differences that impact human immune response and disease during Cryptococcus neoformans infection. mBio. 2019;10(4). https://doi.org/10.1128/mBio.01440-19.
Boral H, Metin B, Dogen A, Seyedmousavi S, Ilkit M. Overview of selected virulence attributes in Aspergillus fumigatus, Candida albicans, Cryptococcus neoformans, Trichophyton rubrum, and Exophiala dermatitidis. Fungal Genet Biol. 2018;111:92–107. https://doi.org/10.1016/j.fgb.2017.10.008.
Steenbergen JN, Casadevall A. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microbes Infect. 2003;5(7):667–75. https://doi.org/10.1016/s1286-4579(03)00092-3.
Zaragoza O. Multiple disguises for the same party: the concepts of morphogenesis and phenotypic variations in Cryptococcus neoformans. Front Microbiol. 2011;2:181. https://doi.org/10.3389/fmicb.2011.00181.
Chrisman CJ, Albuquerque P, Guimaraes AJ, Nieves E, Casadevall A. Phospholipids trigger Cryptococcus neoformans capsular enlargement during interactions with amoebae and macrophages. PLoS Pathog. 2011;7(5):e1002047. https://doi.org/10.1371/journal.ppat.1002047.
Garcia-Rodas R, de Oliveira HC, Trevijano-Contador N, Zaragoza O. Cryptococcal titan cells: when yeast cells are all grown up. Curr Top Microbiol Immunol. 2019;422:101–20. https://doi.org/10.1007/82_2018_145.
Mukaremera L, Lee KK, Wagener J, Wiesner DL, Gow NAR, Nielsen K. Titan cell production in Cryptococcus neoformans reshapes the cell wall and capsule composition during infection. Cell Surf. 2018;1:15–24. https://doi.org/10.1016/j.tcsw.2017.12.001.
Trevijano-Contador N, de Oliveira HC, Garcia-Rodas R, Rossi SA, Llorente I, Zaballos A, et al. Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog. 2018;14(5):e1007007. https://doi.org/10.1371/journal.ppat.1007007.
Crabtree JN, Okagaki LH, Wiesner DL, Strain AK, Nielsen JN, Nielsen K. Titan cell production enhances the virulence of Cryptococcus neoformans. Infect Immun. 2012;80(11):3776–85. https://doi.org/10.1128/IAI.00507-12.
Garcia-Rodas R, Casadevall A, Rodriguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PLoS One. 2011;6(9):e24485. https://doi.org/10.1371/journal.pone.0024485.
Okagaki LH, Nielsen K. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell. 2012;11(6):820–6. https://doi.org/10.1128/EC.00121-12.
Cordero RJ, Casadevall A. Functions of fungal melanin beyond virulence. Fungal Biol Rev. 2017;31(2):99–112. https://doi.org/10.1016/j.fbr.2016.12.003.
Smith DFQ, Casadevall A. The role of melanin in fungal pathogenesis for animal hosts. Curr Top Microbiol Immunol. 2019;422:1–30. https://doi.org/10.1007/82_2019_173. Role of melanin as a virulence factor and possible functions during interactions with environmental hosts.
Taborda CP, da Silva MB, Nosanchuk JD, Travassos LR. Melanin as a virulence factor of Paracoccidioides brasiliensis and other dimorphic pathogenic fungi: a minireview. Mycopathologia. 2008;165(4–5):331–9. https://doi.org/10.1007/s11046-007-9061-4.
Walker CA, Gomez BL, Mora-Montes HM, Mackenzie KS, Munro CA, Brown AJ, et al. Melanin externalization in Candida albicans depends on cell wall chitin structures. Eukaryot Cell. 2010;9(9):1329–42. https://doi.org/10.1128/EC.00051-10.
Wiesner DL, Specht CA, Lee CK, Smith KD, Mukaremera L, Lee ST, et al. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS Pathog. 2015;11(3):e1004701. https://doi.org/10.1371/journal.ppat.1004701.
Noverr MC, Williamson PR, Fajardo RS, Huffnagle GB. CNLAC1 is required for extrapulmonary dissemination of Cryptococcus neoformans but not pulmonary persistence. Infect Immun. 2004;72(3):1693–9. https://doi.org/10.1128/iai.72.3.1693-1699.2004.
Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013;14(2):R11. https://doi.org/10.1186/gb-2013-14-2-r11.
Sykes DE, Band RN. Polyphenol oxidase produced during encystation of Acanthamoeba castellanii. J Protozool. 1985;32(3):512–7. https://doi.org/10.1111/j.1550-7408.1985.tb04052.x.
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D61. https://doi.org/10.1093/nar/gkw1092.
Narita TB, Koide K, Morita N, Saito T. Dictyostelium hybrid polyketide synthase, SteelyA, produces 4-methyl-5-pentylbenzene-1,3-diol and induces spore maturation. FEMS Microbiol Lett. 2011;319(1):82–7. https://doi.org/10.1111/j.1574-6968.2011.02273.x.
Robert V, Cardinali G, Casadevall A. Distribution and impact of yeast thermal tolerance permissive for mammalian infection. BMC Biol. 2015;13:18. https://doi.org/10.1186/s12915-015-0127-3.
Perfect JR. Cryptococcus neoformans: the yeast that likes it hot. FEMS Yeast Res. 2006;6(4):463–8. https://doi.org/10.1111/j.1567-1364.2006.00051.x.
Bloom ALM, Jin RM, Leipheimer J, Bard JE, Yergeau D, Wohlfert EA, et al. Thermotolerance in the pathogen Cryptococcus neoformans is linked to antigen masking via mRNA decay-dependent reprogramming. Nat Commun. 2019;10(1):4950. https://doi.org/10.1038/s41467-019-12907-x.
Casadevall A, Kontoyiannis DP, Robert V. On the emergence of Candida auris: climate change, azoles, swamps, and birds. mBio. 2019;10(4). https://doi.org/10.1128/mBio.01397-19.
Haynes BC, Skowyra ML, Spencer SJ, Gish SR, Williams M, Held EP, et al. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog. 2011;7(12):e1002411. https://doi.org/10.1371/journal.ppat.1002411.
Zhang S, Hacham M, Panepinto J, Hu G, Shin S, Zhu X, et al. The Hsp70 member, Ssa1, acts as a DNA-binding transcriptional co-activator of laccase in Cryptococcus neoformans. Mol Microbiol. 2006;62(4):1090–101. https://doi.org/10.1111/j.1365-2958.2006.05422.x.
Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell. 2005;16(5):2285–300. https://doi.org/10.1091/mbc.e04-11-0987.
Landell MF, Salton J, Caumo K, Broetto L, Rott MB. Isolation and genotyping of free-living environmental isolates of Acanthamoeba spp. from bromeliads in southern Brazil. Exp Parasitol. 2013;134(3):290–4. https://doi.org/10.1016/j.exppara.2013.03.028.
Andra J, Herbst R, Leippe M. Amoebapores, archaic effector peptides of protozoan origin, are discharged into phagosomes and kill bacteria by permeabilizing their membranes. Dev Comp Immunol. 2003;27(4):291–304. https://doi.org/10.1016/s0145-305x(02)00106-4.
Rambach G, Dum D, Mohsenipour I, Hagleitner M, Wurzner R, Lass-Florl C, et al. Secretion of a fungal protease represents a complement evasion mechanism in cerebral aspergillosis. Mol Immunol. 2010;47(7–8):1438–49. https://doi.org/10.1016/j.molimm.2010.02.010.
Guimaraes AJ, Frases S, Cordero RJ, Nimrichter L, Casadevall A, Nosanchuk JD. Cryptococcus neoformans responds to mannitol by increasing capsule size in vitro and in vivo. Cell Microbiol. 2010;12(6):740–53. https://doi.org/10.1111/j.1462-5822.2010.01430.x.
Fu MS, Coelho C, De Leon-Rodriguez CM, Rossi DCP, Camacho E, Jung EH, et al. Cryptococcus neoformans urease affects the outcome of intracellular pathogenesis by modulating phagolysosomal pH. PLoS Pathog. 2018;14(6):e1007144. https://doi.org/10.1371/journal.ppat.1007144.
Schlam D, Canton J, Carreno M, Kopinski H, Freeman SA, Grinstein S, et al. Gliotoxin suppresses macrophage immune function by subverting phosphatidylinositol 3,4,5-trisphosphate homeostasis. mBio. 2016;7(2):e02242. https://doi.org/10.1128/mBio.02242-15.
Gessler NN, Egorova AS, Belozerskaia TA. Melanin pigments of fungi under extreme environmental conditions (review). Prikl Biokhim Mikrobiol. 2014;50(2):125–34. https://doi.org/10.7868/s0555109914020093.
Samarasinghe H, Aceituno-Caicedo D, Cogliati M, Kwon-Chung KJ, Rickerts V, Velegraki A, et al. Genetic factors and genotype-environment interactions contribute to variation in melanin production in the fungal pathogen Cryptococcus neoformans. Sci Rep. 2018;8(1):9824. https://doi.org/10.1038/s41598-018-27813-3.
Joseph J, Chaurasia S, Sharma S. Case report: corneal Coinfection with fungus and amoeba: report of two patients and literature review. Am J Trop Med Hyg. 2018;99(3):805–8. https://doi.org/10.4269/ajtmh.18-0158.
Hsu HY, Ernst B, Schmidt EJ, Parihar R, Horwood C, Edelstein SL. Laboratory results, epidemiologic features, and outcome analyses of microbial keratitis: a 15-year review from St. Louis. Am J Ophthalmol. 2019;198:54–62. https://doi.org/10.1016/j.ajo.2018.09.032.
Polacheck I, Hearing VJ, Kwon-Chung KJ. Biochemical studies of phenoloxidase and utilization of catecholamines in Cryptococcus neoformans. J Bacteriol. 1982;150(3):1212–20. https://doi.org/10.1128/JB.150.3.1212-1220.1982.
Lee SC, Dickson DW, Casadevall A. Pathology of cryptococcal meningoencephalitis: analysis of 27 patients with pathogenetic implications. Hum Pathol. 1996;27(8):839–47. https://doi.org/10.1016/s0046-8177(96)90459-1.
Nosanchuk JD, Valadon P, Feldmesser M, Casadevall A. Melanization of Cryptococcus neoformans in murine infection. Mol Cell Biol. 1999;19(1):745–50. https://doi.org/10.1128/mcb.19.1.745.
Baig AM, Rana Z, Tariq S, Lalani S, Ahmad HR. Traced on the timeline: discovery of acetylcholine and the components of the human cholinergic system in a primitive unicellular eukaryote Acanthamoeba spp. ACS Chem Neurosci. 2018;9(3):494–504. https://doi.org/10.1021/acschemneuro.7b00254.
Casadevall A, Rosas AL, Nosanchuk JD. Melanin and virulence in Cryptococcus neoformans. Curr Opin Microbiol. 2000;3(4):354–8. https://doi.org/10.1016/s1369-5274(00)00103-x.
Mylonakis E, Casadevall A, Ausubel FM. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 2007;3(7):e101. https://doi.org/10.1371/journal.ppat.0030101.
Siddiqui R, Khan NA. Acanthamoeba is an evolutionary ancestor of macrophages: a myth or reality? Exp Parasitol. 2012;130(2):95–7. https://doi.org/10.1016/j.exppara.2011.11.005. One of the most important reviews highlighting the ancestrality of acanthamoeba to macrophages.
de Souza TK, Soares SS, Benitez LB, Rott MB. Interaction between methicillin-resistant Staphylococcus aureus (MRSA) and Acanthamoeba polyphaga. Curr Microbiol. 2017;74(5):541–9. https://doi.org/10.1007/s00284-017-1196-z.
Gupta N, Samantaray JC, Duggal S, Srivastava V, Dhull CS, Chaudhary U. Acanthamoeba keratitis with Curvularia co-infection. Indian J Med Microbiol. 2010;28(1):67–71. https://doi.org/10.4103/0255-0857.58736.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Marina da Silva Ferreira, Diego de Souza Gonçalves, Elisa Gonçalves Medeiros, José Mauro Peralta and Allan J. Guimarães declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Tropical Mycoses
Rights and permissions
About this article
Cite this article
da Silva Ferreira, M., de Souza Gonçalves, D., Medeiros, E.G. et al. “Feast-Fit-Fist-Feat”: Overview of Free-living Amoeba Interactions with Fungi and Virulence as a Foundation for Success in Battle. Curr Trop Med Rep 8, 18–31 (2021). https://doi.org/10.1007/s40475-020-00220-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40475-020-00220-3