Abstract
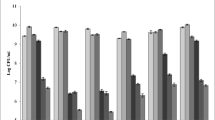
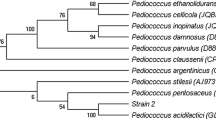
Tannin-degrading bacteria (TDGB) were isolated from the rumen of goats fed oak leaves containing diet. The isolates were screened for their ability to increase in vitro digestibility of oak leaves (Quercus semicarpifolia), and fifteen best isolates were selected for further characterisation. The isolates TDGB 19, 20, 406, 415, 417, 420, 425, 437, 446, and 450 were Gram-positive cocci with medium- and long-chains, and isolates TDGB 409, 428, 430, and 433 were Gram-positive cocco-bacilli of short chain length. All the isolates exhibited tannase activity with the maximum in TDGB 417 (72.0 units; nmol methyl gallate degraded/min/ml) and minimum in TDGB 433 (28.92 units). All the fifteen isolates utilized fructose, galactose, lactose, starch, maltose, glycerol, and mannose as carbon source. The isolates were tested for their tolerance to phenolic monomers like ferulic acid, vanillic acid, coumaric acid, pyrogallol acid, and gallic acid. All the isolates could tolerate the presence of pyrogallol and gallic acid up to 20 mM, but ferulic was toxic at 20 mM level for all the isolates. The isolates were able to degrade tannic acid to pyrogallol except isolates TDGB 409, 428, 430, and 433, which could degrade tannic acid only up to gallic acid after 24 h of incubation as assessed by thin-layer chromatography. None of the isolates degraded tannic acid to resorcinol even after 96 h of incubation. The phylogenetic analysis of the isolates using sequences of PCR amplicons generated by the three sets of primers, two primers sets targeting 16S rRNA gene and one targeting sodA gene for superoxide dismutase enzyme, revealed that the isolates TDGB 7, 19, 20, 406, 417, 446, and 450 formed a close cluster with Streptococcus gallolyticus with 99 to 100 % similarity in all the three phylogenetic trees. There was a maximum of 11 % improvement in vitro true degradability of tannin-rich feed in the presence of live culture of TDGB 406 and can be exploited as a probiotic to improve nutrient utilization of tannin-containing feed given to the livestock.



Similar content being viewed by others
References
Ammar H, Lopez S, Andres S, Ranilla MJ, Bodas R, Gonzalez JS (2008) In vitro digestibility and fermentation kinetics of some browse plants using sheep or goat rumen fluid as the source of inoculum. Anim Feed Sci Technol 147:90–104
Bhat TK, Singh B, Sharma OP (1998) Microbial degradation of tannins–a current perspective. Biodegradation 9:35–343
Bhat TK, Kannan A, Singh B, Sharma OP (2013) Value addition of feed and fodder by alleviating the antinutritional effects of tannins. Agric Res 2:189–206
Brooker JD, O’Donovan LA, Skene I, Clarke K, Blackall L, Muslera P (1994) Streptococcus caprinus sp. nov., a tannin resistant ruminal bacterium from feral goats. Lett Appl Microbiol 18:313–318
Ephraim E, Odenyo A, Ashenafi M (2005) Isolation and characterization of tannin-degrading bacteria from faecal samples of some wild ruminants in Ethiopia. Anim Feed Sci Technol 118:243–253
Goel G, Puniya AK, Aguilar CN, Singh K (2005) Interaction of gut microflora with tannins in feeds (a review). Natuewissenschaften 92:497–503
Goel G, Puniya AK, Singh K (2007) Phenotypic characterization of tannin protein complex degrading bacteria from faeces of goats. Small Rumin Res 69:217–220
Krause DO, Smith WJM, Brooker JD, McSweeney CS (2005) Tolerance mechanisms of streptococci to hydrolysable and condensed tannins. Anim Feed Sci Technol 121:59–75
Krumholz LR, Bryant MP (1986) Syntrophococcus sucromutans sp. nov. gen. nov. uses carbohydrates as electron donors and formate, methoxybenzenoids or Methanobrevibacter as electron acceptor systems. Arch Microbiol 143:31–313
McSweeney CS, Palmer B, Bunch R, Krause DO (2001) In vitro quality assessment of tannin-containing tropical shrub legumes: protein and fibre digestion. Anim Feed Sci Technol 82:227–241
Menke KH, Steingass H (1988) Estimation of the energetic feed value obtained from chemical analysis and gas production using rumen fluid. Anim Res Dev 28:7–55
Mlambo V, Sikosana JLN, Mould FL, Smith T, Owen E, Mueller H (2007) The effectiveness of adapted rumen fluid versus PEG to ferment tannin containing substrates in vitro. Anim Feed Sci Technol 136:128–136
Nelson KE, Pell AN, Schofield P, Zinder S (1995) Isolation and characterization of an anaerobic ruminal bacterium capable of degrading hydrolyzable tannins. Appl Environ Microbiol 61:3293–3298
Nelson KE, Thonney ML, Woolston TK, Zinder SH, Pell AN (1998) Phenotypic and phylogenetic characterization of ruminal tannin-tolerant bacteria. Appl Environ Microbiol 64:3824–3830
Odenyo AA, Osuji PO (1998) Tannin-tolerant ruminal bacteria from East African ruminants. Can J Microbiol 44:905–909
Osawa R, Sly LI (1991) Phenotypic characterization of CO2 requiring strains of Streptococcus bovis from koalas. Appl Environ Microbiol 57:3037–3039
Paul SS, Kamra DN, Sastry VRB, Sahu NP, Kumar A (2003) Effect of phenolic monomers on biomass and hydrolytic enzyme activities of an anaerobic fungus isolated from wild nil gai Baselophus tragocamelus. Lett Appl Microbiol 36:377–381
Pawar MM, Kamra DN, Agarwal N, Chaudhary LC (2014) Effects of essential oils on in vitro methanogenesis and feed fermentation with buffalo rumen liquor. Agric Res 3:67–74
Poyart C, Quesne G, Trieu-Cout P (2002) Taxonomic dissection of the Streptococcus bovis group by analysis of manganese dependent superoxide dismutase gene (sodA) sequences reclassification of Streptococcus infantarius subsp. Coli’ as Streptococcus lutiensis sp. nov. and of Streptococcus bovis biotype 11.2 as Streptococcus pasteurianus sp. nov. Int J Sys Evo Microbiol 52:1247–1255
Sasaki E, Osawa R, Nishitani Y, Whilley RA (2004) Development of a diagnostic PCR assay targeting the Mn-dependent superoxide dismutase gene (sodA) for identification of Streptococcus gallolyticus. J Clin Microbiol 42:1360–1362
Sharma OP, Bhat TK, Singh B (1998) Thin-layer chromatography of gallic acid, methyl gallate, pyrogallol, phloroglucinol, catechol, resorcinol, hydroquinone, catechin, epicatechin, cinnamic acid, p-coumaric acid, ferulic acid and tannic acid. J Chromatogr A 822:167–171
Sharma S, Bhat TK, Dawra RK (2000) A spectrophotometric method for assay of tannase using rhodanine. Anal Biochem 279:85–89
Simpson JM, McCracken VJ, White BA, Gaskins HR, Mackie RI (1999) Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J Microbiol Methods 36:167–179
Singh B, Chaudhary LC, Agarwal N, Kamra DN (2011) Effect of feeding Ficus infectoria leaves on rumen microbial profile and nutrient utilization in goats. Asian-Aust J Anim Sci 24:810–817
Singh B, Chaudhary LC, Agarwal N, Kamra DN (2011) Phenotypic and phylogenetic characterization of tannin degrading/tolerating bacterial isolates from the rumen of goats fed on pakar (Ficus infectoria) leaves. J Appl Anim Res 39:120–124
Singh KM, Pandya PR, Tripathi AK, Patel GR, Parnerkar S, Kothari RK, Joshi CG (2013) Molecular diversity of protozoa in rumen of Indian buffalo (Bubalus bubalis). Agric Res 4:360–366
Skene IK, Brooker JD (1995) Characterization of tannin acylhydrolase activity in the ruminal bacterium Selenomonas ruminantium. Anaerobe 1:321–327
SPSS (2003) Statistical Packages for Social Sciences Version 12.0. SPSS Inc., Chicago, IL, USA
Stahl E (1969) Thin layer chromatography. A laboratory hand book. Springer, New York
Tsai YL, Olson BH (1991) Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol 57:1070–1074
Van Soest PJ, Robertson JB (1988) A laboratory manual for animal science 612. Cornell University, Ithaca
Acknowledgments
The financial assistance provided to senior author in the form of a fellowship by the University Grant Commission, New Delhi and research grant provided by Department of Biotechnology New Delhi, India (Grant No. BT/PR8696/AAQ/01/317/2007) are gratefully acknowledged.
Conflict of interest
There was no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, K., Chaudhary, L.C., Agarwal, N. et al. Isolation and Characterization of Tannin-Degrading Bacteria from the Rumen of Goats Fed Oak (Quercus semicarpifolia) Leaves. Agric Res 3, 377–385 (2014). https://doi.org/10.1007/s40003-014-0121-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40003-014-0121-y