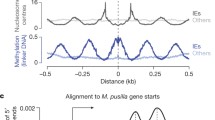
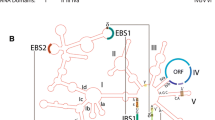
Abstract
Knowing how introns originated should greatly enhance our understanding of the information we carry in our DNA. Gilbert’s suggestion that introns initially arose to facilitate recombination still stands, though not for the reason he gave. Reanney’s alternative, that evolution, from the early “RNA world” to today’s DNA-based world, would require the ability to detect and correct errors by recombination, now seems more likely. Consistent with this, introns are richer than exons in the potential to extrude the stem-loop structures needed for the homology search that can lead to heteroduplex formation and the recognition of base mismatches. In nucleic acid sequences that were unable concomitantly to encode sufficient stem-loop potential, protein-encoding potential was constrained to arise as segments (exons) interrupted by segments rich in stem-loop potential (introns). Thus, sequences with properties that we now deem intronic are likely to have preceded the emergence of exons.





Similar content being viewed by others
References
Battail G (2007) Information theory and error-correcting codes in genetics and biological evolution. In: Barbieri M (ed) Introduction to biosemiotics: the new biological synthesis. Springer, Dordrecht, pp 299–345
Bechtel JM, Wittenschlaeger T, Dwyer T, Song J, Arunachalam S, Ramakrishnan SK, Shepard S, Fedorov A (2008) Genomic mid-range inhomogeneity correlates with an abundance of RNA secondary structures. BMC Genom 9:284. doi:10.1186/1471-2164-9-284
Butler S (1926) The Shrewsbury edition of the works of Samuel Butler. Vol. 20. Jones HF, Bartholomew AT (eds), Jonathan Cape, London
Chargaff E (1951) Structure and function of nucleic acids as cell constituents. Fed Proc 10:654–659
Cock AG, Forsdyke DR (2008) Treasure your exceptions: the science and life of William Bateson. Springer, New York, pp 339–377
Crick F (1971) General model for chromosomes of higher organisms. Nature 234:25–27
Crick F (1979) Split genes and RNA splicing. Science 204:264–271
D’Onofrio G, Mouchiroud D, Aïssani B, Gauter C, Bernardi G (1991) Correlations between the compositional properties of human genes, codon usage, and amino acid composition of proteins. J Mol Evol 32:504–510
Darnell JE (1978) Implications of RNA–RNA splicing in evolution of eukaryotic cells. Science 202:1257–1260
Dawson WK, Yamamoto K (1999) Mean free energy topology for nucleotide sequences of varying composition on secondary structure calculations. J Theor Biol 201:113–140
Doolittle WF (1978) Genes in pieces: were they ever together? Nature 272:581–582
Doolittle RF (1985) The genealogy of some recently evolved vertebrate proteins. Trends Biochem Sci 10:233–237
Faria LCB, Rocha ASL, Kleinschmidt JH, Silva-Filho MC, Bim E, Herai RH, Yamagishi MEB, Palazzo R (2012) Is a genome a codeword of an error-correcting code? PLoS ONE 7(5):e36644. doi:10.1371/journal.pone.0036644
Forsdyke DR (1981) Are introns in-series error detecting sequences? J Theor Biol 93:861–866
Forsdyke DR (1995a) A stem-loop “kissing” model for the initiation of recombination and the origin of introns. Mol Biol Evol 12:949–958
Forsdyke DR (1995b) Conservation of stem-loop potential in introns of snake venom phospholipase A2 genes: an application of FORS-D analysis. Mol Biol Evol 12:1157–1165
Forsdyke DR (1995c) Relative roles of primary sequence and (G + C)% in determining the hierarchy of frequencies of complementary trinucleotide pairs in DNAs of different species. J Mol Evol 41:573–581
Forsdyke DR (1996) Different biological species “broadcast” their DNAs at different (G + C)% “wavelengths.” J Theor Biol 178:405–417
Forsdyke DR (2007a) Calculation of folding energies of single-stranded nucleic acid sequences: conceptual issues. J Theor Biol 248:745–753
Forsdyke DR (2007b) Molecular sex: the importance of base composition rather than homology when nucleic acids hybridize. J Theor Biol 249:325–330
Forsdyke DR (2011a) The interrupted gene. In: Krebs JE, Goldstein ES, Kilpatrick ST (eds) Lewin’s genes X. Jones and Bartlett, Boston, pp. 79–97, 172–175
Forsdyke DR (2011b) Evolutionary bioinformatics, 2nd edn. Springer, New York, pp 249–266
Forsdyke DR (2011c) The selfish gene revisited: reconciliation of Williams–Dawkins and conventional definitions. Biol Theor 5:246–255
Forsdyke DR, Bell SJ (2004) Purine-loading, stem-loops, and Chargaff’s second parity rule: a discussion of the application of elementary principles to early chemical observations. App Bioinf 3:3–8
Gilbert W (1978) Why genes in pieces? Science 271:501
Gilbert W (1981) DNA sequencing and gene structure. Science 214:1305–1312
Gould SJ (1993) Fulfilling the spandrels of world and mind. In: Selzer J (ed) Understanding scientific prose. University of Wisconsin Press, Madison, pp 310–336
Hamming RW (1980) Coding and information theory. Prentice-Hall, Englewood Cliffs
Kleckner N, Weiner BM (1993) Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic and premeiotic cells. Cold Spring Harb Symp Quant Biol 58:553–565
Le S-Y, Maizel JV (1989) A method for assessing the statistical significance of RNA folding. J Theor Biol 138:495–510
Liebovitch LS, Tao Y, Todorov AT, Levine L (1996) Is there an error-correcting code in the base sequence of DNA? Biophys J 71:1539–1544
Matsuo K, Clay O, Kunzler P, Georgiev O, Urbanek P, Schaffner W (1994) Short introns interrupting the Oct-2 POU domain may prevent recombination between POU family genes without interfering with potential POU domain ‘shuffling’ in evolution. Biol Chem Hoppe-Seyler 375:675–683
Morange M (1998) A history of molecular biology. Harvard University Press, Cambridge, MA
Morgan TH (1911) Random segregation versus coupling in Mendelian inheritance. Science 34:384
Orgel LE, Crick FHC, Sapienza C (1980) Selfish DNA. Nature 288:645–646
Penny D, Hoeppner MP, Poole AM, Jeffares DC (2009) An overview of the intron-first theory. J Mol Evol 69:527–540
Pfeifer K, Tilghman SM (1994) Allele-specific gene expression in mammals: the curious case of imprinted RNAs. Genes Devel 8:1867–1874
Prabhu VV (1993) Symmetry observations in long nucleotide sequences. Nucleic Acids Res 21:2797–2800
Reanney DC (1978) Noncoding sequences in adaptive genetics. In: Fox CF, Todaro GJ, Stevens JG (eds) Persistent viruses. Proceedings of the 1978 ICN-UCLA symposium on molecular and cellular biology held in Keystone, Colorado. Academic Press, New York, pp 311–330
Reanney DC (1979) RNA splicing and polynucleotide evolution. Nature 277:598–600
Reanney DC (1984) RNA splicing as an error-screening mechanism. J Theor Biol 110:315–321
Reese V (2002) Mutation repair: a proposed mechanism that would enable complex genomes to better resist mutational entropy, and which suggests a novel function for meiosis. The human behavior and evolution society 14th annual meeting, Rutgers University. Abstracts of presentations to session on “New Developments in Biology,” p 40
Robertson M (1977) Immunoglobulin genes and the immune response. Nature 269:648–650
Rogozin IB, Carmel L, Csuros M, Koonin EV (2012) Origin and evolution of spliceosomal introns. Biol Direct 7:11. doi:10.1186/1745-6150-7-11
Rudner R, Karkas JD, Chargaff E (1968) Separation of B. subtilis DNA into complementary strands. III. Direct analysis. Proc Natl Acad Sci USA 60:921–922
Tiemeier DC, Tilghman SM, Polsky FI, Seidman JG, Leder A, Edgell MH, Leder P (1978) A comparison of two cloned mouse β-globin genes and their surrounding and intervening sequences. Cell 14:237–245
Tomizawa J (1984) Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell 38:861–870
Vinogradov AE (2001) Within-intron correlation with base composition of adjacent exons in different genomes. Gene 276:143–151
Wilkinson DM, Ruxton GD (2012) Understanding selection for long necks in different taxa. Biol Rev 87:616–630
Witkowski JA (1988) The discovery of ‘split’ genes: a scientific revolution. Trends Biochem Sci 13:110–113
Yang S, Yuan Y, Wang L, Li J, Wang W, Liu H, Chen J-Q, Hurst LD, Tian D (2012) Great majority of recombination events in Arabidopsis are gene conversion events. Proc Natl Acad Sci USA 109:20992–20997
Zhang C, Xu S, Wei J-F, Forsdyke DR (2008a) Microsatellites that violate Chargaff’s second parity rule have base order-dependent asymmetries in the folding energies of complementary DNA strands and may not drive speciation. J Theor Biol 254:168–177
Zhang C, Li W-H, Krainer AR, Zhang MQ (2008b) RNA landscape of evolution for optimal exons and intron discrimination. Proc Natl Acad Sci USA 105:5797–5802
Zuker M (1990) Prediction of optimal and suboptimal secondary structure for RNA. Meth Enzymol 183:281–306
Acknowledgments
Queen’s University hosts my intron webpages (http://post.queensu.ca/~forsdyke/introns.htm).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Forsdyke, D.R. Introns First. Biol Theory 7, 196–203 (2013). https://doi.org/10.1007/s13752-013-0090-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13752-013-0090-6