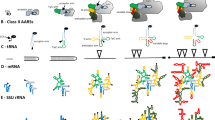
Abstract
We review the introns-first hypothesis a decade after it was first proposed. It is that exons emerged from non-coding regions interspersed between RNA genes in an early RNA world, and is a subcomponent of a more general ‘RNA-continuity’ hypothesis. The latter is that some RNA-based systems, especially in RNA processing, are ‘relics’ that can be traced back either to the RNA world that preceded both DNA and encoded protein synthesis or to the later ribonucleoprotein (RNP) world (before DNA took over the main coding role). RNA-continuity is based on independent evidence—in particular, the relative inefficiency of RNA catalysis compared with protein catalysis—and leads to a wide range of predictions, ranging from the origin of the ribosome, the spliceosome, small nucleolar RNAs, RNases P and MRP, and mRNA, and it is consistent with the wide involvement of RNA-processing and regulation of RNA in modern eukaryotes. While there may still be cause to withhold judgement on intron origins, there is strong evidence against introns being uncommon in the last eukaryotic common ancestor (LECA), and expanding only within extant eukaryotic groups—the ‘very-late’ intron invasion model. Similarly, it is clear that there are selective forces on numbers and positions of introns; their existence may not always be neutral. There is still a range of viable alternatives, including introns first, early, and ‘latish’ (i.e. well established in LECA), and regardless of which is ultimately correct, it pays to separate out various questions and to focus on testing the predictions of sub-theories.





Similar content being viewed by others
References
Bokov K, Sergey V, Steinberg SV (2009) A hierarchical model for evolution of 23S ribosomal RNA. Nature 457:977–980
Brown JWS, Marshall DF, Echeverria M (2008) Intronic noncoding RNAs and splicing. Trends Plant Sci 13:335–342
Carmel L, Rogozin IB, Wolf YI, Koonin EV (2007) Evolutionarily conserved genes preferentially accumulate introns. Genome Res 17:1045–1050
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655
Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA (2002) Selection for short introns in highly expressed genes. Nat Genet 31:415–418
Catania F, Lynch M (2008) Where do introns come from? PLoS Biol 6:e283
Catania F, Gao X, Scofield DG (2009) Endogenous mechanisms for the origins of spliceosomal introns. J Hered 100:591–596
Cavalier-Smith T (2002) The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52:297–354
Cech TR (2009) Crawling out of the RNA world. Cell 136:599–602
Collins LJ, Chen XS (2009) Ancestral RNA: The RNA biology of the eukaryotic ancestor. RNA Biol 6 (in press)
Collins LJ, Penny D (2005) Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol 22:1053–1066
Collins LJ, Penny D (2009) The RNA-infrastructure: dark matter of the eukaryote cell? Trends Genet 25:120–128
Collins LJ, Kurland CG, Biggs P, Penny D (2009) The modern RNP world of eukaryotes. J Hered 100:597–604
Coulombe-Huntington J, Majewski J (2007) Characterization of intron loss events in mammals. Genome Res 17:23–32
Darnell JE, Doolittle WF (1986) Speculations on the early course of evolution. Proc Natl Acad Sci USA 83:1271–1275
Das S (2009) Evolutionary origin and genomic organisation of micro-RNA genes in immunoglobulin Lambda variable region gene family. Mol Biol Evol 26:1179–1189
Davila Lopez M, Rosenblad MA, Samuelsson T (2008) Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucleic Acids Res 36:3001–3010
De Duve C (2007) The origin of eukaryotes: a reappraisal. Nat Rev Genet 8:395–403
De Nooijer S, Holland BR, Penny D (2009) Eukaryote origins: there was no Garden of Eden? PLoS One 4:e5507
Del Campo M, Recinos C, Yanez G, Pomerantz SC, Guymon R, Crain PF, McCloskey JA, Ofengand J (2005) Number, position, and significance of the pseudouridines in the large subunit ribosomal RNA of Haloarcula marismortui and Deinococcus radiodurans. RNA 11:210–219
Dennis PP, Omer A (2005) Small non-coding RNAs in Archaea. Curr Opin Microbiol 8:685–694
Dennis PP, Russell AG, Moniz De Sá M (1997) Formation of the 5′ end pseudoknot in small subunit ribosomal.RNA: involvement of U3-like sequences. RNA 3:337–343
Dennis PP, Omer A, Lowe T (2001) A guided tour: small RNA function in Archaea. Mol Microbiol 40:509–519
Derelle E, Ferraz C, Rombauts S et al (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103:11647–11652
Di Giulio M (2008a) The split genes of Nanoarchaeum equitans are an ancestral character. Gene 421:20–26
Di Giulio M (2008b) Split genes, ancestral genes. In: Wong JT-F, Lazcano A (eds) Prebiotic evolution and astrobiology. Landes Bioscience, Austin
Egel R, Penny D (2007) On the origin of meiosis in eukaryotic evolution: coevolution of meiosis and mitosis from feeble beginnings. In: Egel R, Lankenau D-H (eds) Recombination and meiosis: models, means, evolution. Springer, Berlin, pp 249–288
Eigen M (1992) Steps toward life: a perspective on evolution. Oxford University Press, Oxford
Embley TM, Martin W (2006) Eukaryotic evolution, changes and challenges. Nature 440:623–630
Fedorov A, Fedorova L (2004) Introns: mighty elements from the RNA world. J Mol Evol 59:718–721
Fedorov A, Roy S, Fedorova L, Gilbert W (2003) Mystery of intron gain. Genome Res 13:2236–2241
Forterre P (1995) Thermoreduction, a hypothesis for the origin of prokaryotes. C R Acad Sci Paris Life Sci 318:415–422
Forterre P, Gribaldo S (2007) The origin of modern terrestrial life. HFSP J 1:156–168
Gaffney DJ, Keightley PD (2006) Genomic selective constraints in murid noncoding DNA. PLoS Genet 2:1912–1923
Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A (2009) Rfam: updates to the RNA families database. Nucleic Acids Res 37:D136–D140
Gaspin C, Cavaillé J, Erauso G, Bachellerie J (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J Mol Biol 297:895–906
Gazave E, Marques-Bonet T, Fernando O, Charlesworth B, Navarro A (2007) Patterns and rates of intron divergence between humans and chimpanzees. Genome Biol 8:R21
Gilbert W (1987) The exon theory of genes. Cold Spring Harbor Symp Quant Biol 52:901–905
Gilbert W, de Souza SJ (1999) Introns and the RNA world. In: Gesteland RF, Cech TR, Atkins JF (eds) The RNA world, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 221–231
Goddard MR, Burt A (1999) Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA 96:13880–13885
Guo XY, Wang Y, Keightley PD, Fan LJ (2007) Patterns of selective constraints in noncoding DNA of rice. BMC Evol Biol 7:e208
Halligan DL, Keightley PD (2006) Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res 16:875–884
Hartman A, Fedorov A (2002) The origin of the eukaryotic cell: a genomic investigation. Proc Natl Acad Sci USA 99:1420–1425
Hetzer M, Wurzer G, Schweyen RJ, Mueller MW (1997) Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature 386:417–420
Hickey DA (1992) Evolutionary dynamics of transposable elements in prokaryotes and eukaryotes. Genetica 86:269–274
Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W (2008) RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135:462–474
Irimia M, Penny D, Roy SW (2007a) Coevolution of genomic intron number and splice sites. Trends Genet 23:321–325
Irimia M, Rukov JL, Penny D, Roy SW (2007b) Functional and evolutionary analysis of alternatively spliced genes suggests an early eukaryotic origin of alternative splicing. BMC Evol Biol 7:188
Irimia M, Rukov JL, Penny D, Vinther J, Garcia-Fernandez J, Roy SW (2008) Origin of introns by ‘intronization’ of exonic sequences. Trends Genet 24:378–381
Jeffares DC, Poole AM, Penny D (1998) Relics from the RNA world. J Mol Evol 46:18–36
Jeffares DC, Mourier T, Penny D (2006) The biology of intron gain and loss. Trends Genet 22:16–22
Jeffares DC, Penkett CJ, Bähler J (2008) Rapidly regulated genes are intron poor. Trends Genet 24:375–378
Jurica MS, Moore MJ (2003) Pre-mRNA splicing awash in a sea of proteins. Mol Cell 12:5–14
Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Perlman RE, Roger AJ, Gray MW (2005) The tree of eukaryotes. Trends Ecol Evol 20:670–676
Koonin EV (2006) The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct 1:22
Kurland CG, Collins LJ, Penny D (2006) Genomics and the irreducible nature of eukaryote cells. Science 312:1011–1014
Lane CE, van den Heuvel K, Kozera C et al (2007) Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc Natl Acad Sci USA 104:19908–19913
Lecompte O, Ripp R, Thierry JC, Moras D, Poch O (2002) Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res 30:5382–5390
Lehman N (2003) A case for the extreme antiquity of recombination. J Mol Evol 56:770–777
Lincoln TA, Joyce GF (2009) Self-sustained replication of an RNA enzyme. Science 323:1229–1232
Llopart A, Comeron JM, Brunet FG, Lachaise D, Long M (2002) Intron presence-absence polymorphism in Drosophila driven by positive Darwinian selection. Proc Natl Acad Sci USA 99:8121–8126
Logsdon JM (1998) The recent origin of spliceosomal introns revisited. Curr Opin Genet Dev 8:637–648
Logsdon JM Jr, Tyshenko MG, Dixon C, Jafari D-J, Walker VK, Palmer JD (1995) Seven newly discovered intron positions in the triose-phosphate isomerase gene: evidence for the introns-late theory. Proc Natl Acad Sci USA 92:8507–8511
Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu C (2008) The birth and death of microRNA genes in Drosophila. Nat Genet 40:351–355
Lynch M (2002) Intron evolution as a population-genetic process. Proc Natl Acad Sci USA 99:6118–6123
Lynch M (2007) The origins of genome architecture. Sinauer, Sunderland
Maizels N, Weiner AM (1999) The genomic tag hypothesis: what molecular fossils tell us about the evolution of tRNA. In: Gesteland RF, Cech TR, Atkins JF (eds) The RNA world, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 79–111
Martin W, Russell MJ (2003) On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci 358:59–83
Mosig A, Chen JJ-L, Stadler PF (2007) Homology search with fragmented nucleic acid sequence patterns. In: Algorithms in bioinformatics. Lecture notes in bioinformatics, vol 4645. Springer, Berlin, pp 335–345
Mourier T, Jeffares DC (2003) Eukaryotic intron loss. Science 300:1393
Nedelcu AM (2009) Comparative genomics of phylogenetically diverse unicellular eukaryotes provide new insights into the genetic basis for the evolution of the programmed cell death machinery. J Mol Evol 68:256–268
Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JCD, Andersson DI (2005) Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci USA 102:12112–12116
Niu D-K (2007) Protecting exons from deleterious R-loops: a potential advantage of having introns. Biol Direct 2:11
Ofengand J, Bakin A (1997) Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J Mol Biol 266:246–268
Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP (2000) Homologs of small nucleolar RNAs in Archaea. Science 288:517–522
Omilian AR, Scofield DG, Lynch M (2008) Intron presence-absence polymorphisms in Daphnia. Mol Biol Evol 25:2129–2139
Penny D (2005) An interpretive review of the origin of life research. Biol Philos 20:633–671
Penny D, Collins LJ (2009) Evolutionary genomics leads the way. In: Caetano-Anolles G (ed) Evolutionary genomics and systems biology. Wiley, Hoboken
Penny D, Phillips MJ (2004) The rise of birds and mammals: are microevolutionary processes sufficient for macroevolution. Trends Ecol Evol 19:516–522
Poole AM (2006) Getting from an RNA world to modern cells just got a little easier. BioEssays 28:105–108
Poole AM (2009) Eukaryote evolution: the importance of the stem group. In: Caetano-Anolles G (ed) Evolutionary genomics and systems biology. Wiley, Hoboken
Poole AM, Penny D (2007) Evaluating hypotheses for the origin of eukaryotes. BioEssays 29:74–84
Poole AM, Jeffares DC, Penny D (1998) The path from the RNA world. J Mol Evol 46:1–17
Poole AM, Jeffares DC, Penny D (1999) Prokaryotes, the new kids on the block. BioEssays 21:880–889
Reanney DC (1979) RNA splicing and polynucleotide evolution. Nature 277:598–600
Reanney DC (1987) Genetic error and genome design. Cold Spring Harb Symp Quant Biol 52:751–757
Ren XY, Vorst O, Fiers MWEJ et al (2006) In plants, highly expressed genes are the least compact. Trends Genet 22:528–532
Rodríguez-Trelles F, Tarrío R, Ayala FJ (2006) Origins and evolution of spliceosomal introns. Annu Rev Genet 40:47–76
Rose AB (2008) Intron-mediated regulation of gene expression. Curr Top Microbiol Immunol 326:277–290
Roy SW, Gilbert W (2006) The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet 7:211–221
Roy SW, Irimia M (2009a) Splicing in the eukaryote ancestor: form, function, and dysfunction. Trends Ecol Evol 24:447–455
Roy SW, Irimia M (2009b) Mystery of intron gain: new data and new models. Trends Genet 25:67–73
Roy SW, Penny D (2007) Widespread intron loss suggests retrotransposon activity in ancient apicomplexans. Mol Biol Evol 24:1926–1933
Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Hüttenhofer A (2003) Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31:869–877
Russell AG, Charette JM, Spencer DF, Gray MW (2006) A very early evolutionary emergence of the minor spliceosome. Nature 443:863–866
Santos M, Zintzaras E, Szathmary E (2004) Recombination in primeval genomes: a step forward but still a long leap from maintaining a sizable genome. J Mol Evol 59:507–519
Sashital DG, Cornilescu G, Butcher SE (2004) U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 11:1237–1242
Schoemaker RJ, Gultyaev AP (2006) Computer simulation of chaperone effects of Archaeal C/D box sRNA binding on rRNA folding. Nucleic Acids Res 34:2015–2026
Scofield DG, Lynch M (2008) Evolutionary diversification of the Sm family of RNA-associated proteins. Mol Biol Evol 25:2255–2267
Seetharaman M, Eldho NV, Padgett RA, Dayie KT (2006) Structure of a self-splicing group II intron catalytic effector domain 5: parallels with spliceosomal U6 RNA. RNA 12:235–247
Sharp PA (2009) The centrality of RNA. Cell 136:577–580
Slamovits CH, Keeling PJ (2006) A high density of ancient spliceosomal introns in oxymonad excavates. BMC Evol Biol 6:e34
Stoltzfus A (1999) On the possibility of constructive neutral evolution. J Mol Evol 49:169–181
Sun F-J, Caetano-Anolles G (2008) The origin and evolution of tRNA inferred from phylogenetic analysis of structure. J Mol Evol 66:21–35
Sverdlov AV, Csuros M, Rogozin IB, Koonin EV (2007) A glimpse of a putative pre-intron phase of eukaryotic evolution. Trends Genet 23:105–108
Tanaka H, Kato K, Yamashita E, Sumizawa T, Zhou Y, Yao M, Iwasaki K, Yoshimura M, Tsukihara T (2009) The structure of rat liver vault at 3.5 Angstrom resolution. Science 323:384–388
Tarrio R, Ayala FJ, Rodriguez-Trelles F (2008) Alternative splicing: a missing piece in the puzzle of intron gain. Proc Natl Acad Sci USA 105:7223–7228
Tran E, Zhang X, Lackey L, Maxwell ES (2005) Conserved spacing between the box C/D and C’/D’ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA 11:285–293
Tycowski KT, Aab A, Steitz JA (2004) Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr Biol 14:1985–1995
Valadkhan S, Mohammadi A, Jaladat Y, Geisler S (2009) Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci USA 106:11901–11906
Valentine DL (2007) Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol 5:316–323
Veretnik S, Wills C, Youkharibache P, Valas RE, Philip E, Bourne PE (2009) Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comp Biol 5:e1000315
Wang M, Caetano-Anolles G (2009) The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure 17:66–78
Weiner AM (1993) Messenger-RNA splicing and autocatalytic introns—distant cousins or the products of chemical determinism. Cell 72:161–164
Woodhams MD, Stadler PF, Penny D, Collins LJ (2007) RNase MRP and the RNA processing cascade in the eukaryotic ancestor. BMC Evol Biol 7:S1–S13
Xing Y, Lee C (2007) Relating alternative splicing to proteome complexity and genome evolution. Adv Exp Med Biol 623:36–49
Ying SY, Lin SL (2009) Intron-mediated RNA interference and microRNA biogenesis. Methods Mol Biol 487:387–413
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Penny, D., Hoeppner, M.P., Poole, A.M. et al. An Overview of the Introns-First Theory. J Mol Evol 69, 527–540 (2009). https://doi.org/10.1007/s00239-009-9279-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-009-9279-5