Abstract
Three new lactones, xylanilyticolides A–C (1–3), were isolated from cultures of the actinomycete Promicromonospora xylanilytica YIM 61515. Their structures were elucidated by 1D and 2D NMR spectroscopic data in conjunction with HRESIMS analysis. Compound 1 exhibited potent cytotoxicities against five human cancer cell lines HL-60, A-549, SMMC-7721, MCF-7 and SW480 with the IC50 values of 3.9, 15.2, 11.2, 5.9, and 4.7 μM, respectively.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Actinomycetes are well-known for their ability to produce natural products with structural complexity and diverse biological activities [1, 2]. Salinomycin, isolated from the culture broth of an actinomycete strain of Streptomyces, shows remarkable toxicity for cancer stem cells [3, 4]. Actinomycetes, including some rare genera of actinomycetes [5], are also valuable sources in the discovery of natural antibiotics [6].
Since the genus Promicromonospora was proposed by Krasil’nikov et al. in 1961 [7], there has been only one report of chemical investigation on this genus with discovery of a novel macrocyclic dilactone from Promicromonospora sp. RL26 in 2011 [8]. Herein, the investigation of secondary metabolites from the Promicromonospora xylanilytica YIM 61515 led to the discovery of three new products, xylanilyticolides A–C (1–3) (Fig. 1) [9]. Their structures were elucidated by analyses of 1D and 2D NMR spectroscopic methods. Their cytotoxicities against five human cancer cell lines were evaluated. Report herein are the isolation, structural elucidation and cytotoxicities of them.
2 Results and Discussion
2.1 Structural Elucidation of Xylanilyticolides A–C (1–3)
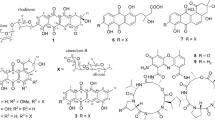
Xylanilyticolide A (1), obtained as colorless oil, had a formula of C26H42O7 deduced by HRESIMS sodium adduct ion peak at m/z 489.2822 [M + Na]+ (calcd. for C26H42O7Na, 489.2823), requiring six degrees of unsaturation. The 1H NMR data displayed three methyl singlets at δH 1.84 (H-9′), 1.52 (H-16), 3.44 (OMe), three methyl doublets at δH 1.54 (H-13, d, J = 6.2 Hz), 1.09 (H-14, d, J = 6.6 Hz), 0.77 (H-15, d, J = 6.6 Hz), and two olefinic protons at δH 5.81 (H-2′, d, J = 15.7 Hz), 7.34 (H-3′, d, J = 15.7 Hz). The 13C NMR and DEPT spectra (Table 1) exhibited signals for 26 carbon atoms, including two carbonyls (δC 165.9, 169.5), six olefinic carbons (δC 115.3, 120.2, 134.4, 134.8, 143.7, 151.1), six methyls (δC 12.8, 13.5, 15.7, 16.1, 20.7, 57.3). The above data coupled with a literature survey indicated that compound 1 was analogous to the reported compound rasfonin, an apoptosis inducer in Ras-dependent cells isolated from the fungus Talaromyces sp. [10]. Comparing with the reported structure, the two distinct differences in compound 1 were the reduction of olefinic bond in C-3 and the substitution of a methoxy in C-4. The 1H-1H COSY correlations between H-3 (δH 2.79, 2.67)/H-4 (δH 3.70), H-4/H-5 (δH 5.29), and HMBC correlation from –OCH3 (δH 3.44) to C-4 (δC 74.1), from H-3 (δH 2.79, 2.67) and H-4 to C-2 (δC 169.5) supported these changes (Fig. 2).
Δ2′ was established as E configuration, which could be confirmed by the coupling constant (see Table 1). Verified by the ROESY correlation between H-3′ (δH 7.34)/H-5′ (δH 5.78), Δ4′ could identified as E configuration. Δ11 was E configuration, which could be identified by the H-12 (δH 5.12)/H-10 (δH 2.00, 1.44) ROESY correlation. From the Newman projection (Fig. 3), the JH-3/H-4 of 3.2 Hz and 5.5 Hz, JH-4/H-5 of 3.2 Hz, JH-6/H-5 of 1.5 Hz as well as H-5/H-6 ROESY correlation were observed, which could confirmed the relative configuration of the lactone ring as 4S*, 5R*, 6R*. However, the stereoconfigurations of C-7, C-9, and C-6′ can’t be determined by the ROESY experiment. The calculated ECD and single crystal X-ray diffraction were also in failure. Therefore, the structure of compound 1 was deduced as xylanilyticolide A.
Compound 2 was obtained as colorless oil. Its molecular formula was assigned as C25H40O6 by HRESIMS ion peak at m/z 459.2723 [M + Na]+ (calcd. for C25H40O6Na, 459.2717), implying the presence of six degrees of unsaturation. The 1H NMR spectrum of 2 illustrated the presence of two methyl singlets at δH 1.54 (H-16) and 1.87 (H-9′); two oxygenated methylenes at δH 3.59/3.49 (H-8′), 3.53 (H-10′); four olefinic protons at δH 5.87 (H-2′), 7.34 (H-3′), 5.84 (H-5′), 5.15 (H-12); and two oxygenated methines at δH 4.79 (H-5), 5.06 (H-6). Combined 13C NMR data (Table 1) and HSQC spectrum indicated the presence of five methyls, seven methylenes, five saturated methines, four olefinic methines, two fully substituted carbons (δC 135.2 and 134.7), and two ketone carbons (δC 176.8 and 167.0). The 1H and 13C chemical shifts of compound 2 (Table 1) were highly similar to those of 1, except the δ-lactone moiety in 1 was replaced by a γ-lactone moiety in 2, which could be verified by the HMBC correlation between H-5 (δH 4.79)/C-2 (δC 176.8) and 1H-1H COSY correlations of H-6 (δH 5.06)/H-5 (δH 4.79), H-5 (δH 4.79)/H-4 (δH 2.38, 1.98), H-4 (δH 2.38, 1.98)/H-3 (δH 2.53,2.47) of compound 2 (Fig. 2). The E/Z configurations of the double bonds in compound 2 were deduced as the same with those of compound 1 by ROESY correlations of H-9′ with H-6′ and H-10′. However, the stereochemistry of C-7, C-9, and C-6′ can’t be established currently. The structure of compound 2 was, therefore, identified as xylanilyticolide B.
The molecular of compound 3 was established by HRESIMS ion peak at m/z 427.2817 [M + Na]+ (calcd. for C25H40O4Na, 427.2819). Detailed analyses of 1H and 13C NMR data of compound 3 revealed that compound 3 had the same skeleton with that of compound 2. The main difference was that C-8′ and C-10′ in compound 3 was two methyl carbons rather than two hydroxymethyl groups in 2. Detailed analysis of 2D NMR data suggested that other parts of 3 were the same to those of 2. Hence the structure of 3 was elucidated as xylanilyticolide C.
2.2 Cytotoxicity Assays
Compounds 1–3 were evaluated for their cytotoxicity activity utilizing an MTS cytotoxicity assay in vitro (DDP was used as a positive control). Compound 1 showed potent inhibitory activity against HL-60, A-549, SMMC-7721, MCF-7 and SW480 cells lines with the IC50 values of 3.9, 15.2, 11.2, 5.9, 4.7 μM, respectively (Table 2), while compounds 2 and 3 showed no significant cytotoxicity activity under the concentration of 40 μM.
3 Experimental
3.1 General Experimental Procedures
Optical rotations were recorded on a JASCO P-1020 digital polari-meter (Horiba, Kyoto, Japan). UV/Vis spectra were obtained using a Shimadzu UV2401PC spectrometer (Shimadzu, Kyoto, Japan). 1D and 2D NMR spectra were measured on a Bruker Avance III 600 MHz spectrometers (Bruker Biospin GmbH, Karlsruhe, Germany). HRESIMS were recorded on an Agilent 6200 Q-TOF MS system (Agilent Technologies, Santa Clara, CA, USA). Silica gel (200–300 mesh, Qingdao Haiyang Chemical Co., Ltd., P. R. China) was used for chromatography column. Size exclusion chromatography was performed on Sephadex LH-20 (GE, Healthcare) columns. MPLC separation was conducted on a Biotage Isolera One using a Spherical C18 column. Semi-preparative HPLC was performed on an Agilent 1260 liquid chromatography system equipped with Zorbax SB-C18 columns (9.4 mm × 150 mm, particle size 5 μm).
3.2 Actinomycete Materials and Culture Condition
The strain was identified as P. xylanilytica YIM 61515 by Prof. Shen Qin of Yunnan University. A voucher strain (HFC: 20160920-4A) was deposited at the School of Pharmaceutical Sciences, South-Central University for Nationalities, China.
The P. xylanilytica YIM 61515 strain was cultured in seed medium (glucose 20 g, peptone 2 g, yeast extract 2 g, soluble starch 5 g, K2HPO4 0.5 g, MgSO4 0.5 g, NaCl 4 g, CaCO3 2 g in 1 L of deionized water, the pH was adjusted to 7.8 before autoclaving) at 25 °C with shaking at 150 rpm for 7 days. Then inoculated the seed culture into 50 Erlenmeyer flasks (500 mL), previously sterilized by autoclaving, each containing 100 g of rice and 100 mL of distilled water. All flasks were incubated at 25 °C for 70 days.
3.3 Extraction and Isolation
Each flask added 200 mL of EtOAc, and the culture was homogenized. The suspension was extracted repeatedly with EtOAc (4 × 10 L), and the organic layer was evaporated to dryness under vacuum to yield a crude extract (49 g), which was fractionated by MPLC with a stepwise gradient of MeOH/H2O (v/v 10:90, 40:60, 70:30, 90:10, 100:0) to give nine fractions (Fr. A–J).
Fr. D (1.87 g) was subjected to 200–300 mesh silica gel column with stepwise elution petroleum ether–EtOAc (8:1–2:1) to obtain nine sub-fractions (D1–D9) based on TLC analyses. The third fraction Fr. D3 was purified by semi-preparative HPLC using a gradient elution (MeCN/H2O 50:50 2.5 mL/min) to give compound 1 (4.4 mg, colorless oil).
Fr. E (508 mg) was separated on Sephadex LH-20 (MeOH) to give four fractions (E1–E4), and compound 2 (3.9 mg) was obtained from Fr. E2 (66.9 mg) which purified with semi-preparative HPLC using an isocratic elution (MeCN/H2O 55:45, 3 mL/min).
Fr. F (1.9 g) was subjected to 200–300 mesh silica gel column eluting with petroleum ether–EtOAc 4:1 to give 5 fractions (F1–F5). Fr. F3 (98.4 mg) was subjected to semi-prep HPLC with isocratic elution (MeCN/H2O 90:10, 3 mL/min) to give compound 3 (0.8 mg).
-
Xylanilyticolide A (1) colorless oil, [α] 24D 8.2 (c 0.11, MeOH); UV (MeOH) λ max(log ε) 202 (3.90), 269 (4.31) nm; 1H and 13C NMR data, see Table 1. HRESIMS m/z 489.2822 [M + Na]+ (calcd. for C26H42O7Na, 489.2823).
-
Xylanilyticolide B (2) colorless oil, [α] 24D 1.7 (c 0.13, MeOH); UV (MeOH) λ max(log ε) 202 (3.85), 268 (4.31) nm; 1H and 13C NMR data, see Table 1. HRESIMS m/z 459.2723 [M + Na]+ (calcd. for C25H40O6Na, 459.2717).
-
Xylanilyticolide C (3) colorless oil, [α] 24D 4.5 (c 0.07, MeOH); UV (MeOH) λ max(log ε) 202 (3.15), 262 (3.77), 544 (1.35) nm; 1H and 13C NMR data, see Table 1. HRESIMS m/z 427.2817 [M + Na]+ (calcd. for C25H40O4Na, 427.2819).
3.4 Cytotoxicity
Five human cancer cell lines, breast cancer SK-BR-3, hepatocellular carcinoma SMMC-7721, human myeloid leukemia HL-60, pancreatic cancer PANC-1, and lung cancer A-549 were used. Cells were cultured in RPMI-1640 or in DMEM medium (Hyclone, Logan, UT, USA), supplemented with 10% fetal bovine serum (Hyclone, USA) in 5% CO2 at 37 °C. The cytotoxicity assay was performed according to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) method in 96-well microplates [11]. Briefly, 100 µL of adherent cells were seeded into each well of 96-well cell culture plates and allowed to adhere for 12 h before addition of test compounds, while suspended cells were seeded just before drug addition with initial density of 1 × 105 cells/mL. Each tumor cell line was exposed to the test compound at concentrations of 0.0625, 0.32, 1.6, 8, and 40 μM in triplicates for 48 h, and all tests were done in twice with cisplatin (Sigma, Los Angeles, USA) as a positive control. After compound treatment, cell viability was detected and a cell growth curve was graphed. IC50 values were calculated by Reed and Muench’s method [12].
References
U.R. Abdelmohsen, K. Bayer, U. Hentschel, Nat. Prod. Rep. 31, 381–399 (2014)
O. Genilloud, Nat. Prod. Rep. 34, 1203–1232 (2017)
P.B. Gupta, T.T. Onder, G. Jiang, K. Tao, C. Kuperwasser, R.A. Weinberg, E.S. Lander, Cell 138, 645–659 (2009)
Y. Miyazaki, M. Shibuya, H. Sugawara, O. Kawaguchi, C. Hirose, J. Nagatsu, S. Esumi, J. Antibiot. 27, 814–821 (1974)
L. Lazzarini, G. Cavaletti, F. Toppo, Marinelli. Antonie Van Leeuwenhoek 78, 399–405 (2000)
J. Clardy, M.A. Fischbach, C.T. Walsh, Nat. Biotechnol. 24, 1541–1550 (2006)
N.A. Krasil’nikov, L.V. Kalakutskii, N.F. Kirillova, Bull. Acad. Sci. USSR Ser. Biol. 1, 107–112 (1961)
M. Izumikawa, M. Takagi, K. Shin-Ya, J. Antibiot. 64, 689–691 (2011)
S. Qin, J.H. Jiang, H.P. Klenk, W.Y. Zhu, G.Z. Zhao, L.X. Zhao, S.K. Tang, L.H. Xu, W.J. Li, Int. J. Syst. Evol. Micr. 62, 84–89 (2012)
H. Fujimoto, Y. Okamoto, E. Sone, S. Maeda, K. Akiyama, M. Ishibashi, Chem. Pharm. Bull. 53, 923–929 (2005)
T. Mosmann, J. Immunol. Methods 65, 55–63 (1983)
L.J. Reed, H. Muench, Am. J. Hyg. 27, 493–497 (1938)
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (81561148013, 21502239), the Key Projects of Technological Innovation of Hubei Province (No. 2016ACA138), and the Fundamental Research Funds for the Central University, South-Central University for Nationalities (CZZ17006, CZQ17008). The authors thank Analytical & Measuring Centre, South-Central University for Nationalities, for the NMR measurements.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, ZX., Qin, S., Xu, LH. et al. Xylanilyticolides A–C, Three New Compounds from Cultures of the Actinomycete Promicromonospora xylanilytica YIM 61515. Nat. Prod. Bioprospect. 8, 91–95 (2018). https://doi.org/10.1007/s13659-018-0154-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13659-018-0154-1