Abstract
A detailed energy-resolved study of the fragmentation reactions of protonated histidine-containing peptides and their b2 ions has been undertaken. Density functional theory calculations were utilized to predict how the fragmentation reactions occur so that we might discern why the mass spectra demonstrated particular energy dependencies. We compare our results to the current literature and to synthetic b2 ion standards. We show that the position of the His residue does affect the identity of the subsequent b2 ion (diketopiperazine versus oxazolone versus lactam) and that energy-resolved CID can distinguish these isomeric products based on their fragmentation energetics. The histidine side chain facilitates every major transformation except trans-cis isomerization of the first amide bond, a necessary prerequisite to diketopiperazine b2 ion formation. Despite this lack of catalyzation, trans-cis isomerization is predicted to be facile. Concomitantly, the subsequent amide bond cleavage reaction is rate-limiting.

Similar content being viewed by others
Introduction
Collision-induced dissociation (CID) of protonated or multiply protonated peptides frequently is used to provide sequence information [1–3]. In favorable cases, fragmentation occurs chiefly by cleavage of the various amide bonds [4] to give a series of bn and ym ions [5, 6] representing, respectively, the N-terminal and C-terminal fragment ions. Thus, cleavage of an amide bond potentially produces a bn ion, a ym ion, or both (for a peptide of n + m residues long) depending on the particular gas-phase chemistries in play. Usually it is these series of bn and ym ions (for a peptide of n + m residues long) that provide the most significant sequence information. However, if these series of fragment ions are incomplete or ambiguous, difficulties may arise in determining the amino acid sequence of the peptide. Consequently, there has been considerable activity in exploring the factors that influence the fragmentation reactions observed, including the structures of the fragment ions formed. Both the reactions observed and the fragment ion structures are expected to be influenced by the amino acid residues in the peptide and the sequence of these residues [4] as this alters the gas-phase structures that are populated at a given level of activation.
It has been established [7, 8] that ym ions are protonated amino acids (y1) or protonated truncated peptides (ym), although the prediction as to which ym ions will be observed is not straightforward. Initially it was proposed [5, 6] that bn ions were substituted acylium ions. However, a number of studies [9–11] have shown that simple b1 ions (α-aminoacylium ions) are unstable and exothermically eliminate CO to form the appropriate iminium ion. Consequently, b1 ions are rarely observed unless some additional means of stabilization is present [11]. Larger bn (n ≥ 2) ions are extensively observed in protonated peptide CID mass spectra. This observation suggests that some interaction within the larger bn ions has occurred to stabilize the ion, an obvious possibility being cyclization to form a structure distinct from the acylium ion structure.
To take a generic b2 ion as an example, there are two possible cyclization reactions, Scheme 1. The first involves cyclization by nucleophilic attack by the adjacent carbonyl group as the amide bond cleaves, resulting in formation of a cyclic protonated oxazolone as shown in Scheme 1b. Extensive tandem MS studies, H/D exchange studies, and theoretical studies of small simple bn ions [12–21] have provided strong evidence for the oxazolone structure, which has also been supported by a number of infrared multiphoton dissociation (IRMPD) studies [22–26] of smaller bn ions.
(a) Nomenclature utilized for protonation sites; (b) generic oxazolone-forming b2 ion pathway; (c) generic diketopiperazine-forming b2 ion pathway; (d) lactam b2 ion pathway. Green dotted lines indicate amide bond stereochemistry is correct for production of a particular product b2 ion. Note that the site of protonation in (b) and (c) can also be on the side chain if this is a basic site
An alternative cyclization involves nucleophilic attack of the N-terminal amine group on the carbonyl function as the amide bond is breaking as illustrated in Scheme 1c. For the b2 case, this results in formation of a protonated diketopiperazine (cyclic dipeptide). Suhai and Paizs [4] have pointed out that for the b2 case, such a cyclization involves a trans-cis isomerization of the amide bond, which is not being broken, an isomerization which has a significant energy barrier. However, there are a number of cases where protonated diketopiperazine formation appears to have been observed for b2 ions. In early work, O’Hair and co-workers [27] observed that the His-Gly and GlyHis b2 ions gave CID product ion mass spectra identical to that of protonated cyclo-(GlyHis) indicating formation of the protonated diketopiperazine structure (Scheme 1c) for the GlyHis and HisGly b2 ions. Recent nuclear magnetic resonance (NMR) data on samples collected from MS/MS of [GlyHisGly + H]+ also support the diketopiperazine assignment, although with the caveat that oxazolone structures are discriminated against with this method as it requires re-dissolving the products prior to purification and NMR analysis [28]. Calculations predict that the diketopiperazine structure with the proton on the imidazole side chain was the most stable species. More recently, Wysocki and co-workers [29, 30] have reported that the His-Ala b2 ion is a mixture of diketopiperazine and oxazolone structures under their experimental conditions. It appears that the His residue catalyzes the trans-cis isomerization, which is necessary prior to diketopiperazine formation. A study [30] of analogues of His showed that the location and accessibility of the histidine π-nitrogen or an amine nitrogen on an aliphatic side chain were necessary for this isomerization to occur. Wysocki and co-workers [31] also have reported that the His-Pro b2 ion likewise has a diketopiperazine structure. This unusual behavior by histidine-containing peptides was also identified in statistical comparisons of doubly protonated peptide spectra performed in the Zubarev group [32]. Histidine in position 2 had the greatest influence of those residues examined on whether Class I or Class II spectra were observed. On this basis, Zubarev et al. [32] hypothesized that doubly protonated peptides with histidine as the second residue in the sequence would produce Class I spectra that contained protonated diketopiperazine rather than protonated oxazolone b2 ion structures. Based on IRMPD studies, Polfer and co-workers [33] have reported that the b2 ion derived by water loss from protonated GlyArg-OH has a diketopiperazine structure whereas that derived from protonated H-ArgGly-OH is a mixture of diketopiperazine and oxazolone structures. Presumably, the difficulty in proton mobilization here [4, 34–36] enables the trans-cis isomerization reaction to occur, making the diketopiperazine competitive. A third possibility exists for peptides with histidine or other potentially nucleophilic side chain in position 2 (Scheme 1d). If the nucleophilic attack on the second amide carbonyl carbon occurs from an imidazole nitrogen of the histidine side chain, a lactam structure will be formed. To complicate matters even further, isomerization of the putative oxazolone product b2 ion structure to this lactam is also potentially possible [37].
In the present work, we initially studied the fragmentation reactions of protonated PheHis-OMe and HisPhe-NH2 and their b2 ions. The reasons for this are: (1) these systems offer a simplified model for investigating proton mobility in doubly protonated peptides with C-terminal proton sequestration, (2) the ongoing debate on b2 ion structure, and (3) interest in the practical details of how the fragmentation mechanisms of these reactions practically occur. We have studied the fragmentation reactions of protonated cyclo-(HisPhe), the synthetic equivalent of gas-phase generated protonated diketopiperazine, and its product ions. In addition, following initial review, we expanded the work to include the related sequences PheHis-NH2, HisPhe-OMe, PheHisAla, and HisPheAla. This enabled a more thorough comparison of the effect of leaving group on the resulting product ion distributions by providing a short series of systematically increasing gas-phase basicity (methanol < ammonia < alanine).
Experimental
Experimental work was carried out using an electrospray/quadrupole/time-of-flight (QqToF) mass spectrometer (QStarXL; SCIEX, Concord, Canada) and a Bruker MaXis plus (Billerica, MA, USA) quadrupole time-of-flight mass spectrometer. The product ion mass spectra for [M + H]+ ions were obtained by mass-selecting the appropriate ion with the quadrupole CID in the collision cell followed by product analysis by the ToF analyser. Data were collected as a function of collision energy. The studies of the fragment ions involved quasi-MS3 experiments. In this approach, CID in the interface region (QStarXL) or between the two ion funnels (MaXis plus) produced fragment ions with those of interest being selected by the quadrupole mass analyzer for CID and mass analysis in the usual fashion. Breakdown graphs expressing the relative fragment ion signals as a function of collision energy were obtained for all species studied.
Ionization was by electrospray (ESI) with the sample at micromolar concentrations in 1:1 CH3OH:1% aqueous formic acid introduced into the source at a flow rate of 10 ųL.min–1 (QStarXL). For the MaXis plus experiments, micromolar acetonitrile/water/formic acid (50/50/0.1%) solutions were utilized at a flow rate of 3 ųL.min–1. Nitrogen was used as nebulizing, drying gas, and as collision gas in both instruments. Energy-resolved breakdown curves for the various analytes were found to be very similar between the two instruments for the same laboratory collision energies. The compounds cyclo-(His-Phe), PheHis-OMe, and HisPhe-NH2 were obtained from Bachem Biosciences (King of Prussia, PA, USA). HisPhe-OMe, PheHis-NH2, HisPheAla, and PheHisAla were obtained from GenSript (Piscataway, NJ, USA). All peptides were used as received.
Theoretical Methods
Standard density functional theory calculations B3LYP [38–40] and M06-2X [41, 42] with the 6-31+g(d,p) basis set were performed with the Gaussian’09 [43] suite of programs. Minima were characterized by harmonic frequency calculations to identify local energy minima (all real frequencies) and transition structures (one imaginary frequency). Multiple transition structures (TSs) were investigated for each reaction. Intrinsic reaction coordinate (IRC) calculations were run for all barriers to determine which minima the TSs connected and thus define the detailed reaction pathway. These consisted of up to 18 steps in each direction along the reaction coordinate. The final structures on both the product and reactant sides of the IRC were then optimized with small incremental steps to identify the connecting minima with confidence.
Results and Discussion
Histidine-Containing Protonated Dipeptide Fragmentation Chemistry
Fragmentation of protonated Phe-His-OMe produces substantial b2 and y1 ions followed by His immonium (m/z 110) and a1 ions. The a1-y1 products become increasingly competitive at higher collision energies consistent with this being an entropically favorable process (loose TS, generation of three gaseous species from the molecular ion rather than two for the other reactions). This is generally consistent with the computed TS energies at both levels of theory [Table 1 for M06-2X/6-31+G(d,p) and Table S1 for B3LYP/6-31+G(d,p)] with the B3LYP modeling making the relative thresholds more similar than M06-2X. Which of the multiple b2 ion-forming pathways is most likely based on these calculations?
Our calculations show the M06-2X/6-31+G(d,p) barriers to ester, C(O)–O, bond cleavage to generate protontated oxazolone and diketopiperazine b2 ion structures as 160 and 141 kJ mol–1, respectively, whereas the B3LYP barriers are essentially identical (162 and 158 kJ mol–1). The C(O)–O bond cleavage barrier is rate-limiting for the protonated diketopiperazine M06-2X/6-31+G(d,p) potential energy surface; trans-cis isomerization requires at least 128.5 kJ mol–1 to generate the cis conformers necessary for the subsequent reaction. This scenario agrees with the predictions of Paizs and Suhai [44] in that amide isomerization reaction may be quite demanding. Practically, catalyzation of the trans-cis isomerization by the histidine side chain does not appear to be a prerequisite for isomerization as we located multiple amide bond rotational pathways that achieved this with the histidine side chain H-bonded elsewhere (Figure 1a, Scheme S2). The subsequent methanol loss reaction occurs in a concerted manner consistent with the unstable nature of the ester oxygen as a protonation site (O3 in Table 1); proton transfer from the histidine side chain to the C-terminal ester oxygen and cleavage of the carbonyl carbon to ester oxygen bond occur consecutively in a single complex motion. The TS is displayed in Figure 1b. The analogous oxazolone-forming pathway also involves the concerted proton transfer and carbonyl carbon to ester oxygen bond cleavage (Figure 1c, Scheme S1). In contrast, neither means of formation of the lactam b2 ion structure (Scheme S3) is predicted to be energetically competitive with protonated oxazolone or diketopiperazine-forming reactions by either level of theory (Table 1, Table S1). Consequently, the protonated diketopiperazine structure is predicted to be the predominant b2 ion structure formed.
Upon collisional activation, the protonated peptide with the reversed amino acid sequence, [His-Phe-NH2 + H]+, initially produced b2 ions followed by abundant a2 and a1 ions. At higher collision energies, these ions began to fragment, leading to the a2-CO-NH3 fragment at m/z 212 (Figure S1). A low abundance peak at m/z 138 corresponding to a b1 ion is also observed fleetingly at intermediate collision energies (dark blue line, Figure S1). The calculated energetics of the [His-Phe-NH2 + H]+ minima and fragment ion-producing reactions are summarized in Table 2 for M06-2X/6-31+G(d,p) and Table S2 for B3LYP/6-31+G(d,p). The obvious outlier in viewing the calculated barriers in light of the experimental data is that of the transition structure to b1 ion production (Figure 2a, Scheme S4), which appears to be inconsistent with our experimental findings as it is lower than the lowest energy b2-NH3 TS. This contradiction is resolved by examining the energies of the separated products (Scheme S4, Table 2), which clearly show that the b1-y1 reaction is product-limited and substantially more energetically demanding (ΔE el + ZPE = 207.3 kJ mol– 1) than any of the b2-NH3 pathways.
For the experimentally predominant b2 ion product, our calculations predict trans-cis isomerization to be facile as it requires over 30 kJ mol–1 less than either protontated oxazolone or diketopiperazine b2 ion formation (Table 2, Figure 2, Schemes S5 and S6). Consequently, the amide bond cleavage is predicted to be the rate-determining step in b2 ion formation for [HisPhe-NH2 + H]+. Despite this, the diketopiperazine b2 ion formation TS barrier is much higher than the oxazolone pathway at both levels of theory. Thus an oxazolone b2 ion is predicted to result from [HisPhe-NH2 + H]+ fragmentation (Figure 2b). We utilize pseudo-MS3 fragmentation of the b2 ion peak in each system to test the predicted fragmentation chemistry. Protonated synthetic cyclo-(HisPhe) was also examined for comparison.
Analysis of b2 Ion and Protonated Cyclo-(HisPhe) Fragmentation
The fragmentation reactions of protonated cyclo-(HisPhe) and the b2 ions (m/z 285) derived from Phe-His-OMe and His-Phe-NH2 have been studied in detail. In addition, the fragmentation reactions of the [M + H – CO]+ ion derived from cyclo-(HisPhe) and the a2 ions derived from protonated PheHis-OMe, HisPhe-NH2 have also been studied in detail. The results for the protonated diketopiperazine cyclo-(HisPhe) are discussed in comparison with the detailed experimental and computational studies published concurrently by Armentrout and Clarke [45], Siu and co-workers [46], and Bythell et al. [47]. Energy-resolved fragmentation of b2 ions has been shown as a means of distinguishing oxazolone from diketopiperazine structures based on the substantially higher barriers to fragmentation present for the protonated diketopiperazine structure and the resulting product ion distributions this provides [45, 47]. On this basis, it should be possible to tell the structures apart in the present case, provided that the presence of the basic histidine residue does not obscure differences in structure.
Figure 3 presents the breakdown graphs obtained on the Bruker MaXis instrument for the b2 ions generated with the original sequences PheHis-OMe (top panel) and HisPhe-NH2 (middle panel), as well as protonated cyclo-(HisPhe) (bottom panel). Similar differences are observed in the breakdown graphs obtained with the QStarXL over a wider collision energy range as shown in Figures S8–S10 of the Supporting Information. What is immediately clear is that there are differences between the b2 ions with the original sequences PheHis (top panel) and HisPhe (middle panel). The latter requires far less energy to fragment than the former and produces abundant a2 and a1 ions as well as the phenylalanine immonium ion with initial onset of fragmentation being ~5 eV laboratory collision energy. Furthermore, the spectrum generated from the b2 ion produced from the protonated PheHis-OMe is very similar to that of protonated cyclo-(HisPhe). This provides strong evidence that the major structure of the b2 ions derived from protonated PheHis-OMe is a protonated diketopiperazine and, thus, the alternate sequence-generated, HisPhe, b2 ion structure is predominantly protonated oxazolone. Our calculations strongly support the alternate sequence-generated, HisPhe, b2 ion being an oxazolone structure too; the energy threshold is more than 20 kJ mol–1 lower for the oxazolone pathway (and this pathway being relatively entropically favorable too, Table 2, Table S2). We cannot entirely rule out the possibility that there is a small contribution from an oxazolone structure in the population of b2 ions generated from protonated PheHis-OMe, but this would likely be very minor based on the experimental results. This is reasonably consistent with our M06-2X findings, which place the protonated diketopiperazine threshold energy ~6 kJ mol–1 lower. The only caveat is why the difference is not larger. At very high ion ‘temperature’, the diketopiperazine b2-MeOH TS becomes more energetically demanding because of the highly unfavorable relative entropy of the diketopiperazine b2-MeOH transition structure (–44.4 J mol–1). Consequently, the oxazolone b2-MeOH TS, which is less entropically hindered, is likely to become more competitive at higher ion temperature, assuming any precursor is still present.
We then calculated the barriers to carbon monoxide loss from the various protonated diketopiperazine and oxazolone forms to determine if this initial fragmentation reaction provided evidence for/against our structural assignments. Decarbonylation of the putative PheHis sequence b2 oxazolone can occur either directly or following isomerization to the lactam structure. The indirect pathway (Scheme 2a) requires less energy to initiate. Isomerization of the b2 oxazolone to form the lactam is facile as it requires only 39.0 kJ mol–1 to initiate (Figure 4a). Subsequent expulsion of the CO from the lactam then requires at least 128.7 kJ mol–1 (Figure 4b). We found that the HisPhe sequence b2 oxazolone has a relatively low energy TS (Figure 4c) corresponding to direct decarbonylation of the protonated oxazolone structure. This requires at least 132.9 kJ mol–1 [ΔE el + ZPE, M06-2X/6-31+G(d,p)] to access (Scheme 2b). For comparison, we also calculated the barrier to the b2 → a1 reaction, which we recently studied for a series of oxazolone b2 ions [48]. This reaction requires substantially more energy, ΔE el + ZPE = 192.3 kJ mol– 1. How do these relatively low barriers compare with fragmentation reactions of [cyclo-(HisPhe) + H]+, the protonated diketopiperazine predicted as the product of [PheHis-OMe + H]+ methanol loss?
Lowest energy decarbonylation reactions of b2 ions (a) PheHis b2 oxazolone ion isomerization to lactam, then CO expulsion; (b) HisPhe b2 oxazolone ion pathway; (c) the complicated, concerted [cyclo(HisPhe`) + H]+, diketopiperazine b2-a2 pathway. We are aware that the a2 ions generated in these pathways may cyclize and/or isomerize subsequently
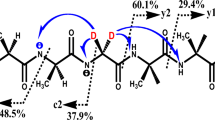
Decarbonylation of the protonated diketopiperazine b2 ion is far more energetically demanding than any of these processes. The lowest energy pathway to fragmentation of this structure has a lot in common with the b2-X fragmentation pathways of the precursor ions (X=NH3 or MeOH). The fragmentation again follows a complicated, concerted mechanism relying on local proton mobility (Scheme 2c). It begins from a low energy His ring-protonated conformer with charge-solvation provided by the adjacent His carbonyl oxygen and additional hydrogen bonding between the phenylaniline ring and its amide hydrogen. Proton transfer from the histidine side chain to the phenylaniline amide nitrogen is followed by elongation of, then cleavage of that amide bond to release CO. The deprotonated histidine side chain effectively guides the CO molecule out of the amide bond after protonating the amide nitrogen by forming a very short-lived, lactam-like transition structure, but with elongated bonds (2.67 Å for the TS; Figure 4d) between the carbonyl carbon and the deprotonated ring-nitrogen. This reaction requires at least 256.7 kJ mol–1 to be active. Numerous alternate pathways were attempted, including ones similar to those described for the protonated cylic peptides, cyclo(GlyGly), cyclo(AlaAla), and cyclo(PheLeu) [45–47], but these were even more energetically costly. Nevertheless, these calculations are consistent with the experimental data displayed in Figure 3 with the b2 ions in the top and bottom panels requiring substantially higher energy collisions in order to initiate significant fragmentation.
Does the C-Terminus Affect the b2 Ion Structures Produced?
We agree with a reviewers comment that we should have also looked at the sequences HisPhe-OMe and PheHis-NH2 experimentally. Consequently, we have had these peptides synthesized along with HisPheAla and PheHisAla to provide a more detailed investigation of leaving group effects on threshold energy and, thus, product ion structure(s). Additionally, we have completed M06-2X/6-31+G(d,p) potential energy surface and TS calculations for these new systems to provide a short series for comparison. These are summarized in Table 3.
These calculations predict the cleavage of the HisPhe–X bond is affected by the nature of the leaving group. This affect is relatively consistent for both the oxazolone and diketopiperazine-forming pathways and follows an approximately linear trend, where increasing gas-phase basicity of the leaving group [49] is correlated with lower HisPhe–X bond cleavage barrier. The diketopiperazine b 2 -X TS is consistently substantially more energetically demanding than the preceding trans → cis amide bond isomerization reaction and also the oxazolone-forming pathway. Conversely, the trans → cis amide bond isomerization reaction barrier is correlated with gas-phase basicity of the C-terminal residue/modification, indicating that a more basic C-terminus is less effective at stabilizing the isomerization reaction. Our calculations predict the oxazolone product ion should predominate in all three cases. Our b 2 ion MS3 experiments (Figure S2) reveal a more complex picture. While the b 2 ions generated from [HisPhe-NH2 + H]+ and [HisPheAla + H]+ show very similar breakdown graphs, which are consistent with the oxazolone structure, the b 2 ions generated from [HisPhe-OMe + H]+ have a different population (Figure S2, top panel). These provide intermediate fragmentation characteristics that show lower prevalence of the a2 ion product and higher collision energy being necessary to achieve over 50% dissociation (~18 eV versus ~12 eV collisions), indicating that a relatively significant population of diketopiperazine ions is present in addition to the oxazolone ions. One possible explanation for this apparent discrepancy is that the kinetic shift associated with the oxazolone b2-X TS may be significantly larger than for the other two cases, thereby enabling more consecutive fragmentation of the oxazolone product to occur at the same time as some formation of the diketopiperazine isomer also occurs. The potential depletion of the oxazolone product would thus lead to a remaining population with an higher diketopiperazine composition. The substantially larger initial b2-X barriers for the [HisPhe-OMe + H]+ form supports this potential explanation.
The situation when histidine in the second residue is predicted to be a lot more complicated. The gas-phase basicity of the leaving group is correlated with lower PheHis-X bond cleavage barrier for the oxazolone pathway, but not for either the diketopiperazine or lactam congeners. As described previously, the diketopiperazine b 2 -X pathway is supported experimentally and theoretically for the C-terminal methoxy ester form. In contrast, the [PheHis-NH2 + H]+ form narrowly predicts the oxazolone pathway as most likely to be active (Table 3, Figure S3), followed by the lactam (+5.1 kJ mol–1), and then the diketopiperazine (+8.5 kJ mol–1). The significantly improved H-bonding available to stabilize the transition structures when NH3 is the leaving group rather than methanol, reduces the barriers here. The [PheHisAla + H]+ system has reduced relative stabilization to the amide-terminated form, as the additional bulk of the leaving group limits the proximity of the H-bonding possible. In the oxazolone and diketopiperazine pathways, this is compensated for in part by the C-terminal carbonyl H-bonding to the N-terminus leading to reduced relative barriers. This stabilization is not available to the lactam TS, so this is relatively less favorable (Figure S3). Overall, the oxazolone and diketopiperazine (+4.1 kJ mol–1) pathways are very similar energetically and entropically. Despite this, at the minimum voltage difference between the ion funnels (Bruker MaXis) necessary to generate b 2 ion signal, our subsequent MS3 experiments on the b 2 ion structures generated from [PheHisAla + H]+ produced breakdown graphs similar to the oxazolone (Figure S4). In contrast, the breakdown graph of the b 2 ions generated from [PheHis-NH2 + H]+ does show evidence of a mixture (Figure S4 top right panel) with fragmentation behavior that is intermediate between those of solely protonated oxazolone or cyclo-(HisPhe). For example, Figure S4 indicates 50% dissociation at ~18 eV collision energy for b 2 ions generated from [PheHis-NH2 + H]+ as opposed to ~22 eV for [cyclo-(HisPhe) + H]+ and b 2 ions generated from [PheHis-OMe + H]+. Based on the M06-2X/6-31+G(d,p) calculations, it is unclear why there is so much protonated diketopiperazine present in this b 2 ion population generated from [PheHis-NH2 + H]+. Nevertheless, it is clear that the nature of the second residue and the leaving group do significantly affect the TS energies and, thus, the resulting product ion structure distributions.
Analysis of a2 Ion and [Cyclo-(HisPhe)-CO]+ Ion Fragmentation
As Figures S5–S7 show, m/z 110 (His immonium ion) and m/z 212 are the major fragmentation products of the [M + H – CO]+ ion irrespective of whether this was generated from an oxazolone of diketopiperazine structure. The m/z 212 fragment corresponds to loss of CO + NH3. Similar products [47, 50, 51] have been observed and studied with theory previously. Eventual formation of iminium ions involves initial generation of a proton-bound complex of two imines. The fragmentation of a2 ions to form iminium ions by way of a proton-bound complex of two imines was first proposed by Siu and co-workers [52] for the GlyGly a2 ion. Support for this pathway was provided by a more detailed study [53] of the fragmentation of a variety of a2 ions, where it was shown that the imine with the greater proton affinity produced the most abundant iminium ion. The His imine has a greater proton affinity (~983 kJ mol–1) than the Phe imine (~929 kJ mol–1), so should overwhelmingly win the battle to keep the proton and, as a result, be much more pronounced in the product ion mass spectrum.
The breakdown graph for the a2 ion (m/z 257) derived from protonated HisPhe-NH2 is presented in Figure S6. The graph differs from the graphs for the other m/z 257 ions (Figures S5 and S7) in showing a distinct signal at m/z 138, the His b1 ion. This b1 ion readily fragments by loss of CO to give m/z 110, suggesting multiple means of generation for the histidine immonium ion. For the GlyGly a2 ion derived from triglycine IRMPD studies and theoretical calculations [54, 55] have shown an N1-protonated 4-imidazolidinone. More recent spectroscopic and theoretical studies [56] of the TyrGly a2 ion showed that in addition to this structure, non-cyclic isomers can be formed if a suitable means of catalyzing proton transfer across the ring is present. Similar results were obtained by Siu and co-workers [57]. In principle, either the histidine or the phenylaniline side chains can do this, thereby lowering the barrier. Stein and co-workers [58] have shown that the Thr side chain is also capable of catalyzing this isomerization. One should note that many of these structures are identical to ones formable from the protonated diketopiperazine. As usual, we cannot eliminate the possibility that there may be a minor route to m/z 110 through the diketopiperazine structure (if present). A more detailed theoretical analysis of the interplay of these a2 ion chemistries will follow elsewhere.
Conclusions
A detailed study of the reactions necessary to form and then fragment b2 ions in an environment of local proton mobility has been undertaken. The results show that the PheHis b2 ion can exist largely as either protonated diketopiperazine or oxazolone (or even lactam, in the case of [PheHis-NH2 + H]+) under our experimental conditions, and that the ion population is a function of the leaving group. Based on our limited series, the less basic the leaving group (methanol < NH3 < alanine), the higher the oxazolone ion population. This is indicated by lower collision energy being necessary to fragment 50% of the precursor ions and diagnostic fragment ions relative presence. In contrast, the HisPhe b2 ion exists largely as a protonated oxazolone. Here, the exception is once again the methoxy form, which shows evidence of the diketopiperazine form too. The reactions necessary to generate these ions rely heavily on local proton mobility primarily facilitated by the histidine side chain. The exception to this appears to be trans-cis isomerization of the first amide bond, which does not require direct involvement of the histidine side chain in the present cases. This reaction was facile for all systems examined with the subsequent –X bond cleavage reaction being rate-limiting.
We also show that the prediction of Armentrout and Clarke [45] that oxazolone and diketopiperazine structures should be discernable based on their energy-resolved fragmentation holds. Based on our experiments and calculations, oxazolone fragmentation is substantially more facile. The fragmentation chemistry of a2 ions derived from protonated PheHis-OMe and HisPhe-NH2, and protonated cyclo-(HisPhe), is a lot more similar than the b2 ion case. Nevertheless differences in the structures or, more likely, the distribution of structures generated from the preceding collisional activation are apparent at lower collisional energies, consistent with the premise that how the ions are made can have a noticeable effect on the product ion population distributions [59–64].
References
Larsen, M.R., Roepstorff, P.: Mass spectrometric identification of proteins and characterization of their post-translational modifications in proteome analysis. Fresenius J. Anal. Chem. 366, 677–690 (2000)
Aebersold, R., Goodlett, D.R.: Mass spectrometry in proteomics. Chem. Rev. 101, 269–295 (2001)
Medzihradszky, K.F.: Peptide sequence analysis. Methods Enzymol. 402, 209–244 (2005)
Paizs, B., Suhai, S.: Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 24, 508–548 (2005)
Roepstorff, P., Fohlman, J.: Proposals for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11, 601 (1984)
Biemann, K.: Contributions of mass spectrometry to peptide and protein structure. Biomed. Environ. Mass Spectrom. 16, 99–111 (1988)
Mueller, D.R., Eckersley, M., Richter, W.: Hydrogen transfer reactions in the formation of “Y + 2” sequence ions from protonated peptides. Org. Mass Spectrom. 23, 217–222 (1988)
Cordero, M.M., Houser, J.J., Wesdemiotis, C.: The neutral products formed during backbone cleavage of protonated peptides in tandem mass spectrometry. Anal. Chem. 65, 1594–1601 (1993)
Van Dongen, W.D., Heerma, W., Haverkamp, J., de Koster, C.G.: The B1 fragment from protonated glycine is electrostatically-bound ion/molecule of CH2=NH2 + and CO. Rapid Commun. Mass Spectrom. 10, 1237–1239 (1996)
Rodriquez, C.F., Vuckovic, A.H., Milburn, R.K., Hopkinson, A.C.: Destabilized carbocations. A comparison of C2H4NS and C2H4NO potential energy surface. J. Mol. Struct. (Theochem) 404, 112–125 (1997)
Bythell, B.J., Csonka, I.P., Suhai, S., Barofsky, D.F., Paizs, B.: Gas-phase structure and fragmentation pathways of singly-protonated peptides with N-terminal arginine. J. Phys. Chem. B 114, 15092–15105 (2010)
Yalcin, T., Khouw, C., Csizmadia, I.G., Peterson, M.R., Harrison, A.G.: Why are b-ions stable species in peptide mass spectra? J. Am. Soc. Mass Spectrom. 6, 1165–1174 (1995)
Yalcin, T., Csizmadia, I.G., Peterson, M.R., Harrison, A.G.: The structures and fragmentation of Bn (n ≥ 3) ions in peptide mass spectra. J. Am. Soc. Mass Spectrom. 7, 233–242 (1996)
Nold, M.J., Wesdemiotis, C., Yalcin, T., Harrison, A.G.: Amide bond dissociation in protonated peptides. Structures of the N-terminal ionic and neutral fragments. Int. J. Mass Spectrom. Ion Proc. 164, 137–153 (1997)
Paizs, B., Lendvay, G., Vékey, K., Suhai, S.: Formation of b2 + ions from protonated peptides. Rapid Commun. Mass Spectrom. 13, 523–533 (1999)
Reid, G.E., Simpson, R.J., O’Hair, R.A.J.: Probing the fragmentation reactions of protonated glycine oligomers via multistage mass spectrometry and gas phase ion molecule hydrogen/deuterium exchange. Int. J. Mass Spectrom. 190/191, 209–230 (1999)
Harrison, A.G., Csizmadia, I.G., Tang, T.-H.: Structures and fragmentation of b2 ions in peptide mass spectra. J. Am. Soc. Mass Spectrom. 11, 427–436 (2000)
Rodriquez, C.F., Shoeib, T., Chu, I.K., Siu, K.W.M., Hopkinson, A.C.: Comparison between protonation, lithiation, and argentination of 5-oxazolones. A study of a key intermediate in gas-phase peptide sequencing. J. Phys. Chem. A 104, 5335–5342 (2000)
Somogyi, Á.: Probing peptide fragment ion structures by combining sustained off-resonance collision-induced dissociation and gas-phase H/D exchange (SORI-HDX) in Fourier transform ion-cyclotron resonance (FT-ICR). J. Am. Soc. Mass Spectrom. 19, 1771–1775 (2008)
Bythell, B.J., Somogyi, Á., Paizs, B.: What is the structure of b2 ions generated from doubly protonated tryptic peptides. J. Am. Soc. Mass Spectrom. 20, 618–624 (2009)
Harrison, A.G.: To b or not to b. The ongoing saga of peptide b ions. Mass Spectrom. Rev. 28, 640–654 (2009)
Polfer, N.C., Oomens, J., Suhai, S., Paizs, B.: Spectroscopic and theoretical evidence for oxazolone ring formation in collision-induced dissociation of peptides. J. Am. Chem. Soc. 127, 17154–17155 (2005)
Polfer, N.C., Oomens, J., Suhai, S., Paizs, B.: Infrared spectroscopy and theoretical studies on gas-phase Leu-enkephalin and its fragments. Direct experimental evidence for the mobile proton. J. Am. Chem. Soc. 129, 5887–5897 (2007)
Yoon, S.-H., Chamot-Rooke, J., Perkins, B.R., Hilderbrand, A.E., Poutsma, J.C., Wysocki, V.H.: IRMPD spectroscopy shows that AGG forms an oxazolone b2 + ion. J. Am. Chem. Soc. 130, 17644–17645 (2008)
Oomens, J., Young, S., Molesworth, S., Van Stipdonk, M.: Spectroscopic evidence for an oxazolone structure of the b2 fragment from protonated trialanine. J. Am. Soc. Mass Spectrom. 20, 334–339 (2009)
Bythell, B.J., Erlekam, U., Paizs, B., Maȋtre, P.: Infrared spectroscopy of fragments derived from tryptic peptides. Chem. Phys. Chem. 10, 883–885 (2009)
Farrugia, J.M., Taverner, T., O’Hair, R.A.J.: Side chain involvement in the fragmentation reactions of the protonated methyl esters of histidine and its peptides. Int. J. Mass Spectrom. 209, 99–112 (2001)
Liu, P., Cooks, R.G., Chen, H.: Structure elucidation of peptide b2 Ions. Angew. Chem. Int. Ed. 54, 1547–1550 (2015)
Perkins, B.R., Chamot-Rooke, J., Yoon, S.H., Gucinski, A.C., Somogyi, Á., Wysocki, V.H.: Evidence of diketopiperazine and oxazolone structures for HA b2 + ion. J. Am. Chem. Soc. 131, 17528–17529 (2009)
Gucinski, A.C., Chamot-Rooke, J., Nicol, E., Somogyi, Á., Wysocki, V.H.: Structural influences on preferential oxazolone versus diketopiperazine b2 + ion formation for histidine analogue-containing peptides. J. Phys. Chem. A 116, 4296–4304 (2012)
Gucinski, A.C., Chamot-Rooke, J., Steinmetz, V., Somogyi, Á., Wysocki, V.H.: Influence of N-terminal residue composition on the structure of proline-containing b2 + ions. J. Phys. Chem. A 117, 1291–1298 (2013)
Savitski, M.M., Fälth, M., Fung, Y.M.E., Adams, C.M., Zubarev, R.A.: Bifurcating fragmentation behavior of gas-phase tryptic peptide dications in collisional activation. J. Am. Soc. Mass Spectrom. 19(12), 1755–1763 (2008)
Zou, S., Oomens, J., Polfer, N.C.: Competition between diketopiperazine and oxazolone formation in water loss products from Arg-Gly and Gly-Arg. Int. J. Mass Spectrom. 316/318, 12–17 (2012)
Burlet, O., Orkiszewski, R.S., Ballard, K.D., Gaskell, S.J.: Charge promotion of low-energy fragmentations of peptide ions. Rapid Commun. Mass Spectrom. 6, 658–662 (1992)
Dongre, A.R., Jones, J.L., Somogyi, A., Wysocki, V.H.: Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: evidence for the mobile proton model. J. Am. Chem. Soc. 118, 8365–8374 (1996)
Bythell, B.J., Suhai, S., Somogyi, A., Paizs, B.: Proton-driven amide bond-cleavage pathways of gas-phase peptide ions lacking mobile protons. J. Am. Chem. Soc. 131, 14057–14065 (2009)
Knapp-Mohammady, M., Young, A., Paizs, B., Harrison, A.: Fragmentation of doubly-protonated Pro-His-Xaa tripeptides: formation of b2 2+ ions. J. Am. Soc. Mass Spectrom. 20, 2135–2143 (2009)
Becke, A.D.: New mixing of Hartree-Fock and local density-functional theories. J. Chem. Phys. 98, 1372–1377 (1993)
Stephens, P.J., Devlin, F.J., Chabalowski, C.F., Frisch, M.J.: Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 98, 11623–11627 (1994)
Lee, C., Yang, W., Parr, R.G.: Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785 (1988)
Zhao, Y., Schultz, N.E., Truhlar, D.G.: Exchange-correlation functional with broad accuracy for metallic and nonmetallic compounds, kinetics, and noncovalent interactions. J. Chem. Phys. 123(16), 161103 (2005)
Zhao, Y., Truhlar, D.G.: The M06 Suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-Class functionals and 12 other functionals. Theor. Chem. Account. 120, 215 (2006)
Frisch, M.J.T., G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G.A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H.P., Izmaylov, A.F., Bloino, J., Zheng, G., Sonnenberg, J.L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery, J.A., Peralta, J. E., Ogliaro, F., Bearpark, M., Heyd, J.J., Brothers, E., Kudin, K.N., Staroverov, V.N., Keith, T., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A., Burant, J.C., Iyengar, S.S., Tomasi, J., Cossi, M., Rega, N. Millam, J.M., Klene, M., Knox, J.E., Cross, J.B., Bakken, V., Adamo, C., Jaramillo, J. Gomperts, R., Stratmann, R.E., Yazyev, O., Austin, A.J., Cammi, R., Pomelli, C., Ochterski, J.W., Martin, R.L., Morokuma, K., Zakrzewski, V.G., Voth, G.A., Salvador, P., Dannenberg, J.J., Dapprich, S., Daniels, A.D., Farkas, O., Foresman, J.B., Ortiz, J.V., Cioslowski, J., Fox, D.J.: Gaussian 09, Revision C.01. Gaussian, Inc., Wallingford, CT (2010)
Paizs, B.S.: Combined quantum chemical and RRKM modeling of the main fragmentation pathways of protonated GGG. I. Cis-trans isomerization around protonated amide bonds. Rapid Commun. Mass Spectrom. 15, 2307–2323 (2002)
Armentrout, P.B., Clark, A.A.: The simplest b2+ ion: determining its structure from its energetics by a direct comparison of the threshold collision-induced dissociation of protonated oxazolone and diketopiperazine. Int. J. Mass Spectrom. 316/318, 182–191 (2012)
Shek, P.Y.I., Lau, J.K.-C., Zhao, J., Grzetic, J., Verkerk, U.H., Oomens, J., Hopkinson, A.C., Siu, K.W.M.: Fragmentations of protonated cyclic-glycylglycine and cyclic-alanylalanine. Int. J. Mass Spectrom. 316/318, 199–205 (2012)
Bythell, B.J., Hendrickson, C.L., Marshall, A.G.: Relative stability of peptide sequence ions generated by tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 23, 644–654 (2012)
Bythell, B.J., Harrison, A.G.: Formation of a1 Ions directly from oxazolone b2 ions: an energy-resolved and computational study. J. Am. Soc. Mass Spectrom. 26, 774–781 (2015)
Hunter, E.P., Lias, S.G.: Evaluated gas phase basicities and proton affinities of molecules: an update. J. Phys. Chem. Ref. Data 27(3), 413–656 (1998)
Bythell, B.J., Molesworth, S., Osburn, S., Cooper, T., Paizs, B., Van Stipdonk, M.: Structure and reactivity of an and an* peptide fragments investigated using isotope labeling, tandem mass spectrometry and density functional theory calculations. J. Am. Soc. Mass Spectrom. 19, 1788–1798 (2008)
Zhang, X.: Gas-phase cleavage of the novel iminium ions [R1-CH+-N=CH-R2↔R1-CH=N+=CH-R2]. An experimental and computational study. J. Mol. Struct. 1056/1057, 219–226 (2014)
El Aribi, H., Rodriquez, C.F., Almeida, D.R.P., Ling, Y., Mak, W.W.-N., Hopkinson, A.C., Siu, K.W.M.: Elucidation of fragmentation mechanisms of protonated peptide ions and their products: a case study on glycylglycylglycine using density functional theory and threshold collision-induced dissociation. J. Am. Chem. Soc. 125, 9229–9236 (2003)
Harrison, A.G., Young, A.B., Schnoelzer, M., Paizs, B.: Formation of iminium ions by fragmentation of a2 ions. Rapid Commun. Mass Spectrom. 18, 1635–1640 (2004)
Verkerk, U.H., Siu, C.-K., Steill, J.D., El Aribi, H., Zhao, J., Rodriquez, C.F., Oomens, J., Hopkinson, A.C., Siu, K.W.M.: a2 ion derived from triglycine: an N1-protonated 4-imidazolidinone. J. Phys. Chem. Lett. 1, 868–872 (2010)
Bythell, B.J., Maître, P., Paizs, B.: Cyclization and rearrangement reactions of an fragment ions of protonated peptides. J. Am. Chem. Soc. 132, 14766–14779 (2010)
Bythell, B.J., Hernandez, O., Steinmetz, V., Paizs, B., Maître, P.: Tyrosine side chain catalyzed proton transfer in the YG a2 ion revealed by theory and IR spectroscopy in the ‘fingerprint’ and X-H (X=C, N, O) stretching region. Int. J. Mass Spectrom. 316/318, 227–234 (2012)
Verkerk, U.H., Zhao, J., Lau, J.K.-C., Lam, T.-W., Hao, Q., Steill, J.D., Siu, C.-K., Oomens, J., Hopkinson, A.C., Siu, K.W.M.: Structures of the a2 ions of Ala-Ala-Ala and Phe-Phe-Phe. Int. J. Mass Spectrom. 330/332, 254–261 (2012)
Simón-Manso, Y., Neta, P., Yang, X., Stein, S.E.: Loss of 45 Da from a2 ions and preferential loss of 48 Da from a2 ions containing methionine in peptide ion tandem mass spectra. J. Am. Soc. Mass Spectrom. 22, 280–289 (2011)
Vachet, R.W., Bishop, B.M., Erickson, B.W., Glish, G.L.: Novel peptide dissociation: gas-phase intramolecular rearrangement of internal amino acid residues. J. Am. Chem. Soc. 119, 5481–5488 (1997)
Tsaprailis, G., Nair, H., Somogyi, Á., Wysocki, V.H., Zhong, W., Futrell, J.H., Summerfield, S.G., Gaskell, S.J.: Influence of secondary structure on the fragmentation of protonated peptides. J. Am. Chem. Soc. 121, 5142–5154 (1999)
Forbes, M.W., Jockusch, R.A., Young, A.B., Harrison, A.G.: Fragmentation of protonated dipeptides containing arginine. Effect of activation method. J. Am. Soc. Mass Spectrom. 18, 1959–1966 (2007)
Prell, J.S., Chang, T.M., Biles, J.A., Berden, G., Oomens, J., Williams, E.R.: Isomer population analysis of gaseous ions from infrared multiple dissociation kinetics. J. Phys. Chem. A 115, 2745–2751 (2011)
Paizs, B., Bythell, B.J., Maître, P.: Rearrangement pathways of the a4 ion of protonated YGGFL characterized by IR spectroscopy and modeling. J. Am. Soc. Mass Spectrom. 23, 664–675 (2012)
Hernandez, O., Paizs, B., Maître, P.: Rearrangement chemistry of an ions probed by IR spectroscopy. Int. J. Mass Spectrom. 377, 173–178 (2015)
Acknowledgments
The authors acknowledge support for this work by start-up funds from the University of Missouri-St. Louis and a College of Arts and Sciences Grant. Calculations were performed locally and at the University of Missouri Bioinformatics Consortium in Columbia, MO and at the University of Missouri-Science and Technology, Rolla, MO. M.T.A. thanks the University of Missouri-St. Louis Saudi Arabia Culture Mission for funding her graduate work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Carissa R. Nelson and Maha T. Abutokaikah contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1911 kb)
Rights and permissions
About this article
Cite this article
Nelson, C.R., Abutokaikah, M.T., Harrison, A.G. et al. Proton Mobility in b2 Ion Formation and Fragmentation Reactions of Histidine-Containing Peptides. J. Am. Soc. Mass Spectrom. 27, 487–497 (2016). https://doi.org/10.1007/s13361-015-1298-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1298-4