Abstract
In this paper, we provide a comprehensive summary of available clear cell renal cell carcinoma (ccRCC) microarray data in the form of meta-analysis of genes differentially regulated in tumors as compared to healthy tissue, using effect size to measure the strength of a relationship between the disease and gene expression. We identified 725 differentially regulated genes, with a number of interesting targets, such as TMEM213, SMIM5, or ATPases: ATP6V0A4 and ATP6V1G3, of which limited or no information is available in terms of their function in ccRCC pathology. Downregulated genes tended to represent pathways related to tissue remodeling, blood clotting, vasodilation, and energy metabolism, while upregulated genes were classified into pathways generally deregulated in cancers: immune system response, inflammatory response, angiogenesis, and apoptosis. One hundred fifteen deregulated genes were included in network analysis, with EGLN3, AP-2, NR3C1, HIF1A, and EPAS1 (gene encoding HIF2-α) as points of functional convergence, but, interestingly, 610 genes failed to join previously identified molecular networks. Furthermore, we validated the expression of 14 top deregulated genes in independent sample set of 32 ccRCC tumors by qPCR and tested if it could serve as a marker of disease progression. We found a correlation of high fucosyltransferase 11 (FUT11) expression with non-symptomatic course of the disease, which suggests that FUT11's expression might be potentially used as a biomarker of disease progression.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) is the most common type of kidney cancer that accounts for 2 % of the world total of all adult malignancies. Its most frequent histological subtype—clear cell renal cell carcinoma (ccRCC)—constitutes 75 % of all kidney tumors with 209,000 new cases per year worldwide [1]. ccRCC arises from the renal cortex, and its lipid- and glycogen-rich cells are “clear” on hematoxylin and eosin staining. ccRCC may have sporadic (>96 %) or familial (<4 %) origin (VHL syndrome) [2]. The majority of ccRCC cases are detected incidentally by ultrasound, CT scan, or MRI, and are diagnosed at the late stage due to asymptomatic course of the disease. The classic symptoms such as hematuria, flank pain, fatigue, and abdominal mass occur rarely and are generally indicative of a more advanced disease [3]. ccRCC is difficult to treat and rarely cured once spread beyond the kidney [4]. If limited to kidneys (40 % of diagnosed cases), the most common curative treatment remains a radical or partial nephrectomy [5]. In advanced stages, targeted (immuno- and antiangiogenic) therapy is introduced. Kinase and mammalian target of rapamycin inhibitors were used in a number of clinical trials and evaluated in the aspect of prognosis improvement [6]. However, their long-term clinical effect remains to be determined and requires larger, homogenous, and well-designed retrospective studies.
In the last decade, a large number of markers has been studied for their prognostic value in ccRCC such as carbonic anhydrase IX, p53, XIAP, HIF1-α, VEGF, and Survivin, but their clinical use remains debatable [7]. The vast majority of research is focused on mechanisms which are deregulated and well described in various cancers such as cell differentiation, angiogenesis, and immunosuppression, but there is a need for identification of targets or pathways unique to ccRCC. A better understanding of molecular pathogenesis of ccRCC is required to direct novel therapeutic intervention of the individual patient and to predict patient's prognosis. Several genomic alterations were suggested to be associated with ccRCC tumorgenesis; however, currently there are no accepted molecular biomarkers to monitor ccRCC development [8].
Molecular markers could be incorporated into future staging systems and hold great promise for more accurate prognoses of ccRCC. Advances in technology, such as gene arrays and high-throughput tissue arrays, make the detection of such markers more visible [3].
However, single microarray studies suffer from several problems: they may report findings not reproducible or not robust to data perturbations, [9–11]. Several meta-analysis techniques have been proposed in the context of microarrays so far; however, a comprehensive framework on how to carry out a meta-analysis of microarray data set emerged only recently [12].
In this paper, we provide the results of a meta-analysis of nine selected ccRCC studies using effect size as a measure of the strength of a relationship between two variables (here: the disease and expression of a gene). The goal of this study was to identify genes that are differentially expressed between ccRCC and normal tissue, to group them according to their function (pathway analysis) and to validate a number of potential biomarkers in a homogenous patient group, well defined with respect to VHL, HIF1A, EPAS1 expression and clinical parameters. In order to test prognostic value of the most deregulated genes, we performed logistic regression analyses of clinical and molecular parameters, and showed an association of high expression of the fucosyltransferase gene (FUT11) with non-symptomatic disease course up to 31 months post-surgery.
Material and methods
Study selection and data set preparation
Twelve Affymetrix studies of biopsy confirmed, primary ccRCC samples with TNM, F grades, or WHO classifications were included. The data, in the FLEO format, were obtained from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) [13] and Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) [14]. There were no technical replicates, no information regarding batch effects or image files. For each study, array density plots, MA plots, Spearman correlation plots, and RNA degradation plots were created to reject low quality arrays. Arrays were normalized using the Robust Multichip Average method [15]. Eight studies fulfilled inclusion criteria and 222 tumor and 85 control samples were subjected to the analysis. For each array type, the probes were mapped to version 14 Unigene gene identifiers (Microarray Lab of the University of Michigan, http://brainarray.mbni.med.umich.edu/brainarray/Database/CustomCDF/) [16].
Estimation of a study-specific differential expression of each gene
Effect size was used to measure differential expression of each gene using the following formula:
with μ 1 is the average signal intensity in tumor samples, μ 2 the average signal intensity in controls, σ g pooled standard deviation, and J a constant. Variance ω g of θ g was used as a weight while combining study-specific estimates of differential expression of each gene into a single effect size value.
Combination of study-specific estimates into a single statistic
All genes that were found in less than four studies were removed. An inverse variance technique was used to combine study-specific effect size values into a weighted average, and, for each gene, the following formula was used (k is the number of studies):
p value of the summary effect size was calculated and adjusted for multiple testing using the FDR method.
Data analysis
Computations were performed using the R software (www.r-project.org) and the BioConductor package (http://www.bioconductor.org/). Output genes were converted to Ensemble and Entrez gene ID formats with SOURCE (http://source.stanford.edu) and Biomart (http://central.biomart.org/) ID converters. With the aid of SOURCE or GeneCards Human Gene Database (http://genecards.org), they were also annotated with location, function, and Gene Ontology terms (http://www.geneontology.org/) (Gene Ontology Consortium). The genes were subject to the Gene Functional Classification tool of the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/home.jsp) [17]. Correlation and logistic regression analyses were performed using IBM SPSS Statistics 21.
Patient material
Tumors were collected from patients from Western Poland who were diagnosed with urological carcinomas. In one case, two tumors were detected (patient 01–068) and both tumors were tested for expression of selected genes. The tissues were histopatologically verified as ccRCC and screened for VHL mutations, promoter methylation, expression of VHL, HIF1A and EPAS1, and LOH (data not shown). The study was approved by the local ethical committee (876/09); only patients who signed written consent were included in the study. Disease progression was defined as local disease recurrence or distant metastasis detected by X-ray and abdominal ultrasound, and/or abdominal and pectoral CT. Follow-up time of the patients differs per case. In general, first follow-up visits were carried out approximately 6 or 12 months post-nephrectomy. For detailed patient characteristics, see Online Resource 1, Table S1. The control samples comprised of nine histopathologically unchanged tissues matched to 9 of 32 tumors tested.
qPCR
Primers were designed using Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast) and Oligo Analyzer 3.1 (http://eu.idtdna.com/analyzer/applications/oligoanalyzer/default.aspx). One microgram of RNA was reversely transcribed using RevertAid™ First Strand cDNA Synthesis Kit with Random Hexamers (Thermo Scientific Fermentas, Waltham, MA, USA), following supplied protocol. All analyses were performed on Eco Real-Time PCR System (Illumina, San Diego, CA, USA) using Maxima™ SYBR Green/ROX qPCR Master Mix (2×) (Thermo Scientific Fermentas), following supplied protocol. Using cDNA from non-histopathologically changed tissues, standard curves were prepared. All analyzed samples were compared to ACTB as a reference gene and non-histopathologically changed tissue as a control, and corrected by reaction efficiency obtained from standard curves. Each measurement was performed in duplicate, in two independent runs. The qPCR results of controls were averaged and used for analysis of all tumor tissues.
Results
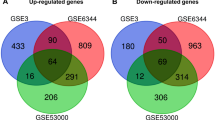
We gathered expression data from eight published microarray studies (Table 1) and performed meta-analysis on a data set derived from 222 tumor and 85 control samples. Seven hundred twenty-five differentially expressed genes were identified for which the summary effect size was lower than −2.5 or greater than 2.5, with FDR less than 0.01 (both cutoffs arbitrarily selected). The top 25 up- and downregulated genes identified in our analysis are listed in Table 2.
First, using GeneCards, we investigated expression patterns of the downregulated genes (Fig. 1). Interestingly, 24 of the top 25 downregulated genes were highly expressed mainly in the kidney, with only one gene, SERPINA5, being highly expressed additionally in other organs. Limited information was available on the expression pattern of FAM3B in any of the listed organs. However, FAM3B, also known as PANcreatic DERived factor (PANDER), has been recently reported to be decreased in gastric cancers with high invasiveness and metastasis [18]. A few of the downregulated genes were described previously (UMOD, KCNJ1, or SERPINA5), but there is a limited information available on the involvement of, for example, TMEM213, SMIM5, or TMEM52B in ccRCC. In general, we observed that downregulated genes tend to represent biological pathways related to tissue remodeling and wound repair, blood clotting, vasodilatation, and energy metabolism (Fig. 2). Genes involved in tissue remodeling and wound repair (e.g., CGN, TUBAL3, EGF, PLG) represent primarily cell adhesion processes such as GAP junction, intercellular channels, extracellular matrix (ECM) remodeling, urokinase-plasminogen activator (PLAU) signaling, and plasmin signaling. The blood clotting pathway is exemplified by genes involved in blood coagulation such as F11, SERPINA5, PROC, and KNG1. The genes taking part in vasodilatation (e.g., XPNPEP2, KNG1, PLG) are involved mainly in peptide hormone signal transduction—bradykinin/kallidin pathway. SREBF2, SCAP, CASR, and NXPH2 represent lipid-associated energy metabolism.
Supporting previously reported data, gene ontology analysis shows a significant clustering of downregulated genes in processes related to ion transport and homeostasis (e.g., cation/anion transport: sodium, potassium, iron), and proper development and function of the kidney nephron, and development of kidney epithelium, renal and urogenital systems (Table 3).
Interestingly, protein products of the downregulated genes were assigned to the three specialized cell compartments: (1) ECM, playing a significant role in the regulation of numerous cellular functions, like cell shape determination, adhesion, migration, proliferation, polarity, differentiation, apoptosis, and wound healing [19, 20]; (2) integral membrane proteins, serving as entry and exit routes for many ions, nutrients, waste products, hormones, drugs, and large molecules (DNA and proteins); (3) membrane vehicles (endosomes and lysosomes) implemented in signal transduction, as well as morphogenetic aspects of normal cell physiology adhesion and migration (Online Resource 1, Table S2).
The 25 top upregulated genes are highly expressed in nearly all examined tissues (Fig. 3), except mitotic CDCA2 and sperm-associated antigen—SPAG4. Majority of the most upregulated genes have been previously described (e.g., HIG2, EGLN3, IKBIP, and VIM), but we also found a few genes with less well-described function in ccRCC, like alpha-(1,3)-fucosyltransferase 11 (FUT11), shown to be expressed in HEK293 cell line, and E3 ubiquitin ligase RNF149 [21].
All upregulated genes were classified into pathways generally deregulated in cancer: immune system response, inflammatory response, DNA damage response, mitogenic signaling, angiogenesis, and apoptosis (Fig. 2). The immune response is represented by alternative and classical complement pathway (e.g., C3, ITGB2, HLA-DRB), antigen presentation by MHC class I and class II genes, HSP60, HSP70, and TLR signaling pathway. Twenty-eight upregulated genes were assigned to inflammatory response pathways such as HSP60, HSP70, TLR, NF-κB, and TNFR1 signaling pathways, TCR and CD28 co-stimulation in activation of NF-κB (e.g., UBC, LY96, TRADD, BID). DNA damage response was represented by a group of inhibitors of apoptosis, which display both anti-apoptotic and pro-survival properties, and their expression can be induced by different cellular stresses such as hypoxia, endoplasmic reticular stress, and DNA damage [22].
The upregulated genes were assigned primarily to the processes of immune response regulation (both positive and negative), cytokine-mediated processes (IFN-γ, cytokine stimulation), and antigen presentation (Table 3). The localization of upregulated gene products was determined mainly as cytoplasmic, but the proteins were also assigned to two additional compartments: cellular membranes and vehicles, supporting the general idea of deregulation of intra- and extracellular signal transduction in ccRCC, similarly to other cancers (Online Resource 1, Table S3).
Network analysis
MetaCore GeneGo program was used to analyze networks of direct interactions between all genes identified in our meta-analysis. Sixty-two differentially regulated genes created a network of interacting genes. The significant interactions between genes (FDR <0.05) are shown in Online Resource 1, Table S4. Our analysis revealed one major network with three distinct central nodes: UBC, AP-2, and GCR-β located centrally and with extensive connections to other genes (Fig. 4). Notably, there were several nodes (plasmin, caspase 1, ENaC, protein C inhibitor, and tissue kallikreins) interconnecting central networks. Inclusion of VHL, HIF1A, EPAS1, and HIF3A in the analysis resulted in addition of the next 53 deregulated genes to the network (e.g., PFKP, HIG2, DARS, HLA-E, JMJD1A). We observed that HIF1A joined the subset of central nodes to create a comprehensive network with others genes (e.g., PFKP, HIG-2, and KNG). EPAS1 was shown to interact with 23 genes (e.g., EGLN3, HLA-E, and VEGFA) (Fig. 4).
Validation of the expression of the most deregulated genes in independent sample set
We set to validate the expression of the seven most down- and the seven most upregulated genes in 32 tumor samples derived from 31 ccRCC patients of Greater Poland. We concentrated on candidate genes not previously reported due to differences in methodology used or not discussed by others: HIG2, NDUFA4L2, EGLN3, FUT11, PRDX4, PFKP, RNF149, TMEM213, HS6ST2, DMRT2, CLDN8, TMEM52B, ATP6V1G3, and ATP6V0A4. First, tumor samples were examined for VHL, HIF1A, and EPAS1 expression. Similarly to previously reported data, we observed 20–50 % reduction of VHL mRNA levels as compared to healthy tissue (data not shown [23]).
All seven genes found as most downregulated in our meta-analysis had decreased expression in all tested tumors. Heparan sulfate 6-O-sulfotransferase 2 (HS6ST2) was downregulated 267-fold on average and detected in 31 (96.88 %) samples (Fig. 5). Average 772-fold downregulation of Doublesex and Mab-3 Related Transcription Factor 2 (DMRT2) was found in 25 (78.12 %) tumors. We detected on average 94-fold lower expression of TMEM52B in 23 (71.88 %) samples. Transmembrane protein 213 (TMEM213) and Claudin 8 (CLDN8) were detected only in three (0.94 %) and four (1.25 %) tumor specimens, with average folds of 1,066 and 226, respectively. The expression of ATP6V0A4 was found in 20 (62.5 %) tumors and was down 1,308 times on average, while the expression of ATP6V1G3 was undetectable in all samples.
The relative expression of hypoxia inducible lipid 2 (HIG2), found upregulated in our meta-analysis, was increased, on average 56-fold, in all tumors tested as compared to the healthy tissue (Fig. 6). The upregulation (on average 122-fold) of mitochondrial NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, 4-like 2 (NDUFA4L2) was found in all tumors, whereas EGL9 homolog 3 (EGLN3) was overexpressed in 31 (96.88 %) samples and 32-fold on average. The mean upregulation of phosphofructokinase (PFKP) and fucosyltransferase 11 (FUT11) was equal to 7.6 and 2.9, respectively (overexpressed in 32 (100 %) and 28 (87.5 %) samples). The expression of Peroxiredoxin 4 (PRDX4) was up in 28 (87.5 %) tumors and increased 1.92-fold on average, while RNF149 was overexpressed approximately 2.26-fold in one tumor specimen. The increased expression of majority of validated upregulated genes (n = 5, except HIG2 and RNF149), correlated with the expression of VHL, HIF1A, and EPAS1, with highest correlation coefficient for FUT11 (0.71), EGLN3 (0.60), PFKP (0.58), and NDUFA4L2 (0.48), and lowest for PRDX4 (0.39), indirectly suggesting their dependence on VHL, HIF1A, and EPAS1. Further, we investigated if the expression of validated genes could be used as a predictor of disease progression using the same group of 31 ccRCC patients. Patient characteristics are shown in Table 4. Due to incomplete clinical data, four patients were excluded from the analysis. First, using forward logistic regression, we found that, out of all genes tested, FUT11's expression was associated with disease progression (p = 0.025, OR = 0.392, 95 % CI = (0.173–0.891)) (ATP6V1G3, TMEM213, and CLDN8 were excluded due to strong downregulation). Combined analysis of clinical and molecular parameters showed that FUT11 remained a significant parameter in the model (p = 0.042, OR = 0.215, 95 % CI = (0.049–0.949)), together with TNM (p = 0.024, OR = 2.379, 95 % CI = (1.124–5.036)) and diabetes (p = 0.047, OR = 0.003, 95 % CI = (0.000–0.924)).
Secondly, we examined the correlation between FUT11 mRNA levels and disease course up to 31 months post-nephrectomy. The patients were divided into two groups: individuals with high FUT11 expression (values greater than the average expression in tumors, n = 14) and low FUT11 expression (n = 14). We observed that majority (12 out of 17) of patients with non-symptomatic disease course displayed high FUT11 expression. We found inverse correlation between the two variables (linear correlation coefficient ρ = −0.51), what, taking under consideration the fact that ccRCC is a complex, multigenic disease, may suggest the importance of FUT11 expression in the ccRCC pathology. In Kaplan–Meier survival analysis, the association between FUT11 expression and non-sympomatic disease course did not reach statistically significant value, most likely to due to relatively small sample set analyzed (see Fig. 7). The results of univariate and multivariate survival analyses with Cox regression model to assess FUT11's input independently of other predictor variables are shown in Table 5.
Discussion
In the present study, we performed a meta-analysis of ccRCC gene expression data derived from public repositories and executed extensive analyses of pathways, processes, and cellular localization of the differentially expressed genes. In our meta-analysis, we implemented the methodology proposed by Ramasamy et al. [12], based on estimation of effect size, in contrast to standard fold change analysis. Effect size measures the overlap of the distributions of signal intensity in cases and controls for each gene, but does not impart how much a gene's expression has changed between cases and controls in terms of fold change. And since effect size does not depend on actual expression values, but on a relationship between them, it is suitable for the analysis of data derived from different experiments, carried out in different conditions and on different platforms.
Using this approach, we found 725 genes deregulated in the tumor tissue. The generated list included genes extensively described in previous reports such as downregulated (UMOD, KNG1, SERPINA5, KCNJ1) and upregulated (EGLN3, VIM, HIG2) [24–27], but it also contained genes of less recognized function in ccRCC pathogenesis, represented by TMEM213 and ATPases: ATP6V1G3, ATP6V0A4, ATP6V0D2.
In line with previous reports, the downregulated genes were mapped to biological processes and pathways essential for proper kidney function (e.g., ion transport and homeostasis) and development (e.g., nephron and kidney epithelium development) [26], whereas the upregulated genes were classified into pathways known to be deregulated in cancer: immune system response, inflammatory response, DNA damage response, angiogenesis, and apoptosis [28].
Network analysis highlighted a few important genes as points of functional convergence, including those recently described in ccRCC: EGLN3 [29], AP-2, NR3C1, kallikreins, and well recognized HIF1A, EPAS1, and genes encoding ubiquitin. Interestingly, only 115 of the 725 deregulated genes were included in the networks, not only supporting the importance of VHL/HIF pathways in ccRCC pathology but also highlighting the significance of additional processes in the development of this disease, as suggested by others [23, 30].
Regression analysis of expression of the validated genes in combination with clinical data showed potential applicability of FUT11 expression as a marker of non-symptomatic disease. Interestingly, we observed correlation of high FUT11 expression with non-symptomatic disease course.
FUT11 belongs to a family of fucosyltransferases—globular type II transmembrane Golgi-resident proteins. Their function is to catalyze the transfer of α-l-fucose from GDP-Fuc onto N- and O-linked glycans, free oligosaccharides, lipids, or directly onto proteins; however, the fucosyltransferase activity has not been confirmed for FUT11 [31]. Fucose, as a constituent of oligosaccharides, is associated with cancer and inflammation [32].
Currently, there is no information available concerning the role of FUT11 in ccRCC, but its upregulation has been detected in additional microarray data sets [33, 34]. FUT11 has also been found upregulated in autosomal dominant polycystic kidney disease expression data [35]. First functional data were provided by Groux-Degroote et al. [33], who showed that IL-6 and IL-8 have stimulatory effect on expression of FUT11, and FUT11 may be involved in the biosynthesis of sialyl-Lewisx and 6-sulfo-sialyl-Lewisx epitopes in the bronchial mucins in inflammatory mucosae of cystic fibrosis patients. Lewis epitopes are crucial for leukocyte homing and extravasation process, thus are essential for lymphocyte maturation and the function of immune system [36]. On the other hand, IL-6 plays an important role in the immune defense mechanism and cell growth and differentiation modulation in numerous malignancies [37]. It has been observed that expression of fucosylated oligosaccharides changes in cancer and inflammation (e.g. [31]), also in ccRCC [38]; hence, detection of FUT11 upregulation in our meta-analysis and in ccRCC tumors may link FUT11 to ccRCC development and progression.
Although our preliminary data suggests involvement of FUT11 in ccRCC progression, our findings require independent validation on additional large sample sets. Further functional studies are needed to acquire more detailed knowledge on the role of this fucosyltransferase in ccRCC development.
References
Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–32.
Pavlovich CP, Schmidt LS. Searching for the hereditary causes of renal-cell carcinoma. Nat Rev Cancer. 2004;4(5):381–93.
Rosette JM, Sternberg CN, van Poppel HP. Renal cell cancer: diagnosis and therapy. London: Springer; 2008.
Lovisolo JA, Casati B, Clerici L, Marafante E, Bono AV, Celato N, et al. Gene expression profiling of renal cell carcinoma: a DNA microarray analysis. Br J Urol Int. 98(1):205–16.
Rathmell WK, Godley PA, Rini BI. Renal cell carcinoma. Curr Opin Oncol. 2005;17(3):261–7.
Pal SK, Figlin RA, Reckamp K. Targeted therapies for non-small lung cancer: an evolving landscape. Mol Cancer Ther. 2010;9(7):1931–44.
Sun M, Shariat SF, Cheng C, Ficarra V, Murai M, Oudard S, et al. Prognostic factors and predictive models in renal cell carcinoma: a contemporary review. Eur Urol. 2011;60(4):644–61.
Rydzanicz M, Wrzesiński T, Bluyssen HA, Wesoly J. Genomics and epigenomics of clear cell renal cell carcinoma: Recent developments and potential applications. Cancer Lett. doi:10.1016/j.canlet.2013.08.006.
MacPherson JI, Sidders B, Wieland S, Zhong J, Targett-Adams P, Lohman V, et al. An integrated transcriptomic and meta-analysis of hepatoma cells reveals factors that influence susceptibility to hcv infection. Plos One. 2011;6(10).
Smith DD, Saetrom P, Snove OJ, Lundberg C, Rivas GE, Glackin C, et al. Meta-analysis of breast cancer microarray studies in conjunction with conserved cis-elements suggest patterns for coordinate regulation. BMC Bioinforma. 2008;9(63).
Anders M, Fehlker M, Wang Q, Wissmann C, Pilarsky C, Kemmner W, et al. Microarray meta-analysis defines global angiogenesis-related gene expression signatures in human carcinomas. Mol Carcinog. 2013;52(1):29–38.
Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. Plos Med. 2008;5(9):1320–32.
Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31(1):68–71.
Barett T, Edgar R. Gene Expression Omnibus (GEO): microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–69.
Irrizary RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64.
Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20).
Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5).
Li Z, Mou H, Wang T, Xue J, Deng B, Qian L, et al. A non-secretory form of FAM3B promotes invasion and metastasis of human colon cancer cells by upregulating Slug expression. Cancer Lett. 2013;328(2):278–84.
Schultz GS, Davidson JM, Kirsner RS, Bornstein P, Herman IM. Dynamic reciprocity in the wound microenvironment. Wound Repair Regen. 2011;19(2):134–48.
Zent R, Pozzi A. Cell–extracellular matrix interactions in cancer. New York: Springer; 2010.
Hong SW, Jin DH, Shin JS, Moon JH, Na YS, Jung KA, et al. Ring Finger Protein 149 is an E3 ubiquitin ligase active on wild-type v-Raf murine sarcoma viral oncogene homolog B1 (BRAF). J Biol Chem. 2012;287(28):24017–25.
Marvin A, Berthelet J, Plenchette S, Dubrez L. The inhibitor of apoptosis (IAPs) in adaptive response to cellular stress. Cells. 2012;1(4):711–37.
Gordan JD, Lal P, Dondeti VR, Letrero R, Parekh KN, Oquendo CE, et al. HIF-α effects on c-Myc distinguish two subtypes of sporadicVHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14(6):435–46.
Xu K, Cui J, Olman V, Yang Q, Puett D, Xu Y. A comparative analysis of gene-expression data of multiple cancer types. Plos One. 2010;5(10).
Togashi A, Katagiri T, Ashida S, Fujioka T, Maruyama O, Wakumoto Y, et al. Hypoxia-inducible protein 2 (HIG2), a novel diagnostic marker for renal cell carcinoma and potential target for molecular therapy. Cancer Res. 2005;65(11):4817–26.
Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, et al. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. Plos One. 2010;5(5).
Wozniak MB, Le Calvez-Kelm F, Abedi-Ardekani B, Byrnes G, Durand G, Carreira C, et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. Plos One. 2013;8(3).
Romaschin AD, Youssef Y, Chow TW, Siu KW, DeSouza LV, Honey RJ, et al. Exploring the pathogenesis of renal cell carcinoma: pathway and bioinformatics analysis of dysregulated genes and proteins. Biol Chem. 2009;390(2):125–35.
Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–3.
Brannon AR, Haake SM, Hacker KE, Pruthi RS, Wallen EM, Nielsen ME, et al. Meta-analysis of clear cell renal cell carcinoma gene expression defines a variant subgroup and identifies gender influences on tumor biology. Eur Urol. 2012;61(2):258–68.
Moriwaki K, Miyoshi E. Fucosylation and gastrointestinal cancer. World J Hepatol. 2010;2(4):151–61.
Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129(1):123–34.
Groux-Degroote S, Krzewinski-Recchi MA, Cazet A, Vincent A, Lehoux S, Lafitte JJ, et al. IL-6 and IL-8 increase the expression of glycosyltransferases and sulfotransferases involved in the biosynthesis of sialylated and/or sulfated Lewisx epitopes in the human bronchial mucosa. Biochem J. 2008;410(1):213–23.
Cifola I, Spinelli R, Beltrame L, Peano C, Fasoli E, Ferrero S, et al. Genome-wide screening of copy number alterations and LOH events in renal cell carcinomas and integration with gene expression profile. Mol Cancer. 2008;7(6).
Song X, Di Giovanni V, He N, Wang K, Ingram A, Rosenblum ND, et al. Systems biology of autosomal dominant polycystic kidney disease (ADPKD): computational identification of gene expression pathways and integrated regulatory networks. Hum Mol Genet. 2009;18(13):2328–43.
Chen HL. Lewis glyco-epitopes: structure, biosynthesis, and functions. Adv Exp Med Biol. 2011;705:53–80.
Guo Y, Xu F, Lu T, Duan Z, Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38(7):904–10.
Borzym-Kluczyk M, Radziejewska I, Darewicz B. Glycosylation of proteins in healthy and pathological human renal tissues. Folia Histochem Cytobiol. 2012;50(4):599–604.
Acknowledgments
We would like to thank the patients for their participation in this study. This work was supported by Foundation for Polish Science, grant “Focus” (3/2008, project leader: J. Wesoły). The qPCR analysis was performed in Genome Analysis Laboratory funded by National Multidisciplinary Laboratory of Functional Nanomaterials NanoFun nr POIG.02.02.00-00-025/09 (Innovative Economy Operational Programme, Priority Axis 2: R&D Infrastructure, Action 2.2: Support of Formation of Common Research Infrastructure of Scientific Units).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 49 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Zodro, E., Jaroszewski, M., Ida, A. et al. FUT11 as a potential biomarker of clear cell renal cell carcinoma progression based on meta-analysis of gene expression data. Tumor Biol. 35, 2607–2617 (2014). https://doi.org/10.1007/s13277-013-1344-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-1344-4