Abstract
Identification of the precise molecular pathways involved in oncogene-induced transformation may help us gain a better understanding of tumor initiation and promotion. Here, we demonstrate that SOX2+ foregut epithelial cells are prone to oncogenic transformation upon mutagenic insults, such as KrasG12D and p53 deletion. GFP-based lineage-tracing experiments indicate that SOX2+ cells are the cells-of-origin of esophagus and stomach hyperplasia. Our observations indicate distinct roles for oncogenic KRAS mutation and P53 deletion. p53 homozygous deletion is required for the acquisition of an invasive potential, and KrasG12D expression, but not p53 deletion, suffices for tumor formation. Global gene expression analysis reveals secreting factors upregulated in the hyperplasia induced by oncogenic KRAS and highlights a crucial role for the CXCR2 pathway in driving hyperplasia. Collectively, the array of genetic models presented here demonstrate that stratified epithelial cells are susceptible to oncogenic insults, which may lead to a better understanding of tumor initiation and aid in the design of new cancer therapeutics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer arises from a progressive accumulation of genetic mutations in proto-oncogenes and tumor suppressor genes (Visvader and Lindeman, 2012; Blanpain and Simons, 2013). For example, the oncogene Kras and the tumor suppressor gene p53 are frequently mutated in a wide range of human cancers (Serrano et al. 1997; Kuilman et al., 2010) and are known to induce tumor initiation in a variety of mouse models (Jackson et al., 2001; Singh et al., 2010).
Abnormal proliferative signals of oncogenic insults including oncogenic KRAS are known to activate a senescent phenotype in cells, presumably designed to prevent the growth of oncogene-transformed cells and to preserve the tumor in a non-aggressive state (Collado and Serrano, 2006). Senescent cells, in turn, secrete large amounts of cytokines and chemokines in a phenomenon known as Senescence-Associated Secretory Phenotype (SASP). Among SASP-related factors, CXC chemokines that bind to CXC chemokine receptor 2 (CXCR2) have been shown to reinforce senescence, which results in growth arrest, further preventing tumor progression (Acosta et al., 2008). However, SASP components can also dangerously stimulate a malignant phenotype and have tumor-promoting responses. Some of the factors secreted by senescent cells such as GROα, CXCL-12 or IL-8 lead to activate proliferation in the surrounding epithelial cells (Krtolica et al., 2001; Coppé et al., 2008). Therefore, the effect of SASP on cell behavior is context-dependent.
Not only is the specific genetic mutation a determining factor for tumor initiation but the cell type from which the tumor originates is also important. Cellular populations that seem to have particularly high tumorigenic potential include adult stem cells (ASCs) and progenitor cells (PCs), which normally play crucial roles in tissue homeostasis and repair (Huels and Sansom, 2015; Sanchez-Danes et al., 2016; Zhu et al., 2016). These cells might be ideal candidates to serve as the cells-of-origin for cancers and as such ASCs/PCs have been intensively studied. However, it still remains to be fully understood which cell population is prone to oncogenic transformation and what kind of oncogenic insults induce tumor initiation from certain ASCs/PCs.
Here, we sought to identify proliferative ASCs/PCs that are the most susceptible to oncogenic mutations. By initially focusing on oncogenic Kras, together with the loss of p53, we found that foregut basal cells that express SOX2 efficiently proliferated to hyperplasia in response to oncogenic mutations. We also revealed distinct roles of oncogenic KRAS and P53 deletion in driving hyperplasia. Furthermore, oncogenic Kras elevated expression of SASP-related chemokines, which contributed to the oncogenic proliferation through a CXCR2-dependent signaling pathway. Taken together, these results suggest that SOX2+ epithelial basal cells in the esophagus and stomach are highly susceptible to oncogenic stimuli. Our findings may help elucidate early events in tumor formation and the cells-of-origin of tumors, which could in turn provide insights towards a better understanding of neoplasia.
RESULTS
Expressing oncogenic Kras and p53 deletion in SOX2+ cells induces hyperplasia in the esophagus and forestomach
To determine which stem cell populations are the most vulnerable to oncogenic transformation, we expressed oncogenic Kras (G12D) and deleted one copy of the p53 gene in dividing cells of the adult mouse. Oncogenic Kras and p53 mutations were chosen because they are frequently observed in a wide range of human cancers (Serrano et al., 1997; Kuilman et al., 2010). We targeted proliferative cell populations using Mcm2-CreER knock-in mice (Mcm2CreER/WT), in which CreER expression is controlled by the Mcm2 promoter. MCM2 is a component of the DNA replication licensing complex and localizes exclusively to proliferating cells. Mcm2 expression is known to be downregulated when homozygous Mcm2-CreER mice (Mcm2CreER/CreER) are used, resulting in the loss of ASCs/PCs and the formation of cancer (likely because of genome instability) (Pruitt et al., 2007). Mcm2CreER/WT mice were bred with mice carrying a loxP-STOP-loxP (LSL)-oncogenic Kras (G12D) (KrasLSL-G12D/WT) and loxP-p53-loxP mice (p53Flox/Flox) (Marino et al., 2000; Jackson et al., 2001). Upon genotyping, we verified and selected mice carrying the appropriate genetic modifications, namely Mcm2CreER/WT; KrasG12D/WT; p53Flox/WT (hereafter referred to as MKPFlox/WT mice). MKPFlox/WT mice allow for the selective induction of KrasG12D expression and the heterozygous deletion of p53 in all dividing cells upon tamoxifen (TAM) administration. These mice also carried an LSL-luciferase (Luc) transgene in the ROSA26 gene locus (ROSALSL-Luc/WT) to allow for the visualization of Cre-expressing cells via bioluminescence imaging (BLI) (Fig. 1A). One month after TAM administration, we performed BLI of MKPFlox/WT mice carrying ROSALSL-Luc/WT and noticed high levels of Luc expression, primarily in digestive tissues, including the small intestine (Fig. 1B). We also observed a prominent hyperplastic forestomach with abnormal proliferation of stratified epithelial layers (Fig. 1B and 1C). We repeated the experiments giving TAM intraperitoneally and the same phenotype was observed (data not shown). We then repeated this experiment using Cre lines restricted to stem cell populations, namely Sox2-CreER (SKPFlox/WT) and Lgr5-CreER (LKPFlox/WT), because SOX2 and LGR5 are known to mark ASC/PC populations in stratified epithelial squamous layers and in lower digestive tracts, respectively (Barker et al., 2007; Arnold et al., 2011). BLI revealed that Luc signals were specifically observed in the esophagus and stomach of SKPFlox/WT mice, whereas LKPFlox/WT mice exhibited strong Luc signals in the duodenum, small intestine, and colon (Fig. S1A), in agreement with previous reports (Feng et al., 2011; Snippert et al., 2014). We did not observe any hyperplasia in animals that lacked the CreER drivers (Fig. S1B).
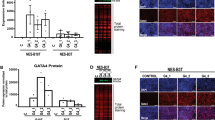
Susceptibility of proliferating cells to oncogenic stimuli. (A) Schematic representation of the genetic strategy for Kras and p53 modifications in MCM2+ cells (MKP mouse model). (B) BLI analysis of Mcm2CreER/WT or MKPFlox/WT 4 weeks post tamoxifen (TAM) administration. Li: Liver; H: Heart; E: Esophagus; St: Stomach; Du: Duodenum; SI: Small intestine; C; Colon; Sp: Spleen; Lu: Lung; K: Kidney; P: Pancreas; B: Brain. (C) H&E on paraffin-embedded sections from Mcm2CreER/WT mice and MKP mice. Scale bars, 100 μm
Although SOX2 is expressed in a broad array of tissues, including lung, trachea, testis, tongue, pituitary gland, eye and brain (Que et al., 2009; Arnold et al., 2011), we did not see any hyperplasia in these tissues in the Sox2-CreER mice (Figs. 2A–C and S2, data not shown). Instead, we observed tissue-specific phenotypes, namely KRAS/P53-driven hyperplasia was generally restricted to the forestomach and esophagus with abnormalities in the glandular stomach. The hyperplasia was observed even in the older (3–4 month old) mice without any difference from the younger ones. We thus focused our attention on SOX2+ cells. SOX2 localizes to basal cells in the esophagus and forestomach, which are known to be progenitor cells with a high proliferative potential (Arnold et al., 2011; Doupe et al., 2012). To characterize hyperplasia in the esophagus and forestomach in more detail, we repeated the SOX2 experiment using a GFP marker (rather than the Luc marker) to allow for immunohistochemistry (IHC)-based lineage tracing of the SOX2+ ASCs/PCs in the esophagus, stomach, and lung (Fig. S3) after TAM administration. We confirmed the appearance of GFP+ cells 1 week after TAM administration in both the esophagus and forestomach (Fig. S4). IHC-based analysis of SKPFlox/WT mice revealed GFP+ cells in the hyperplastic squamous region of the esophagus and forestomach and some of GFP+ cells were positive for KI67, a marker of proliferation (Fig. 2C). Analysis of the abnormalities found in the glandular stomach of SKPFlox/WT mice revealed the presence of high amounts of mucosa, as assessed by Periodic acid-Schiff (PAS) staining (Fig. S5A). The alterations observed in the glandular region, however, were not directly linked to SOX2+ cells because we did not detect a clear increase in the GFP+ population with and without induction of oncogenic activity or a change in the expression pattern of differentiation markers of the glandular stomach proton-pump and gastrin (Fig. S5B). We next asked if oncogenic insults affected the differentiation potential of SOX2+ cells. These GFP+ cells were heterogeneous, with subpopulations expressing markers of undifferentiated (P63) or differentiated (CK13 and LORICRIN) cell types (Fig. S6), suggesting that the KRAS/P53 oncogenic stimulus does not affect the ability of these cells to differentiate, in contrast to what has been observed following Sox2 overexpression (Liu et al., 2013). Previous reports showed that KrasG12D does not seem to be commonly mutated in human esophageal squamous cell carcinoma (ESCC) (Shigaki et al., 2013), although related pathways are often activated (Lin et al., 2014) and this mutation is also observed in the Chinese population (Liu et al., 2011). Therefore, we next examined the effect of PIK3CA (H0147R), which is a mutation associated with ESCC (Lin et al., 2014; Song et al., 2014). Hyperplasia was also observed in the esophagus and forestomach when oncogenic PIK3CA was expressed together with heterozygous p53 deletion (Fig. S7). Together, these results indicate that SOX2+ cells can be the cells-of-origin of forestomach and esophagus hyperplasia and suggest that SOX2+ basal cells in the esophagus and forestomach seem more susceptible to oncogenic stimuli than SOX2+ cells from other tissues in the body, implying tissue-specific vulnerabilities upon oncogenic insults.
Cell susceptibility of foregut epithelial basal cells to oncogenic stimuli. (A) Schematic representation of SKP mouse carrying ROSALSL-GFP for lineage tracing purposes. (B) Stomachs collected from SKPFlox/WT with or without treatment with TAM. (C) Lineage tracing experiment of SOX2+ cells in SKPFlox/WT mice 3 weeks post TAM administration. Co-staining for GFP with KI67, a proliferative marker. Scale bars, 100 μm
Differential impacts of oncogenic KRAS and P53 deletion on gene expression signature
To ascertain whether oncogenic Kras, heterozygous loss of p53, or both were responsible for induced hyperplasia in this context, we analyzed transgenic mice in which Kras and/or p53 were manipulated using different combinations in SOX2+ cells. Upon TAM administration, stomach hyperplasia was only observed in animals that expressed mutant Kras, indicating that KrasG12D expression, but not p53 heterozygous deletion, was sufficient to induce the hyperplastic phenotype. Notably, hyperplasia was observed in almost all SKPFlox/WT mice whereas lower rates were observed in mice carrying only mutant Kras (Fig. 3A), suggesting that deletion of one copy of p53 accelerates tumorigenic proliferation by expanding SOX2+ cells, as supported by our BLI measurements (Fig. 3B) and IHC observations (Fig. S8). To characterize the molecular events that contribute to abnormal proliferation in the presence of oncogenic Kras, we next performed RNA-Sequencing (RNA-Seq) analysis using samples from the forestomach, esophagus, and lungs of Sox2-CreER mice with/without KrasG12D and with/without one copy of the p53 gene (see Fig. 3C). Clustering analysis revealed that gene expression signatures of esophagus and stomach tissue were altered by KrasG12D expression with or without heterozygous p53 deletion. In contrast, these genetic manipulations did not affect gene expression signatures in the lung, where proliferation was not observed. Gene ontology enrichment analysis further indicated a distinct impact of oncogenic KRAS versus P53 deletion (Fig. 3D). Because Kras mutation was sufficient to initiate hyperplasia in SOX2+ cells, we sought to identify specific KRAS target genes. Comparing esophagi and stomachs in which Kras or Kras/p53 were manipulated to controls that did not express Kras (false discovery rate (FDR) < 5%), we identified 13 genes that were upregulated. These included Keratin 17 (Krt17), which is a known marker of malignancy (Du et al., 2013). Of note, some of these KRAS target genes encode secreted factors (Serpine1, Il1b, Cxcl1, Cxcl3, Cxcl5 and Cxcl7) (Fig. 3C and 3E). Importantly, a large fraction of these genes are associated with SASP (Coppe et al., 2008). These genes were upregulated by oncogenic KRAS rather than by P53 modification (Fig. 3E), recapitulating the different impacts of oncogenic KRAS and P53 deletion.
Distinct impacts of oncogenic KRAS and P53 modification. (A) Summary of the incidence of hyperplastic stomach. (B) Ex vivo BLI analysis of tissues from the indicated mice. Li: Liver; H: Heart; E: Esophagus; St: Stomach; Du: Duodenum; SI: Small intestine; C; Colon; Sp: Spleen; Lu: Lung; K: Kidney; P: Pancreas; B: Brain. (C) Heat map showing clustered gene expression profiles in indicated conditions using the results from RNA-Seq. Common genes that are upregulated by oncogenic Kras are extracted between the esophagus and stomach. (D) Gene Ontology enrichment for biological processes in genes significantly regulated by Kras and p53 disruption in each tissue (FDR < 5%, fold-change > 2). (E) Heat map of common upregulated chemokine genes in Fig. 3C
SASP-related factors are involved in oncogenic Kras-mediated cellular proliferation
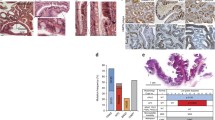
Previous reports have indicated that SASP accelerates the proliferation of tumor cells while inhibiting the proliferation of surrounding wild-type cells (Acosta et al., 2008; Coppe et al., 2008; Kuilman et al., 2008). Therefore we first asked if the CXC chemokines pathway is activated in foregut epithelia. As shown in Fig. 4A, CXCL7, encoded by Cxcl7, which is one of the upregulated SASP-regulated genes, and CXCR2, which is a receptor for the CXC family of chemokines, are expressed in stratified epithelia of the esophagus and forestomach. This led us to examine the effect of CXC chemokines on cell proliferation. For mouse primary esophageal epithelial cells (mpEECs), chemokine treatment accelerated proliferation, highlighting the involvement of these factors in hyperplasia (Fig. 4B). More importantly, chemical inhibition of the CXCR2 signaling pathway with the compound SB225002 (White et al., 1998) in SKPFlox/WT mice (1-week following TAM exposure) resulted in a marked decrease in proliferating cells (BrdU+ cells) and in a thinner hyperplastic layer, to levels comparable to the control mice (Figs. 4C and S9). Analyses of RNA-Seq data from ESCC samples available in public datasets (Tong et al., 2012) showed upregulation of CXC ligands and IL1b (Fig. S10). The ability of CXC ligands and IL1b to enhance tumor effects was also observed in a soft-agar assay utilizing human primary esophageal epithelial cells (Fig. 4D). We next tested CXCR2 inhibitor on human esophageal cell lines: human primary esophageal epithelial cells (hpEECs); non-neoplastic, immortalized esophageal epithelial cells (Het-1A); and ESCC line (OE21). We noticed that CXCR2 inhibitor negatively affected esophageal cell proliferation while not affecting human dermis skin fibroblast (HDF) (Fig. S11), highlighting the importance of CXCR2 in ESCC, consistent with previous report (Wang et al., 2006). Collectively, these data indicate that SASP-related factors play crucial roles in tumorigenesis caused by oncogenic KRAS.
Crucial roles of secretory phenotype on tumor initiation caused by KRAS activation. (A) Expression of CXCL7 and CXCR2 in striated epithelial layers. Scale bars, 100 μm. (B) Effect of chemokines on mouse primary esophageal epithelial cell (mpEEC) proliferation. The isolated esophageal cells were treated with or without recombinant IL1b and CXC ligands (25 ng/mL) for 6 days and then the number of cells was counted. CXCLs: CXCL1, CXCL3, CXCL5 and CXCL7. Data represent the mean with SD (n = 3). *P < 0.01; Student’s t-test. (C) Top, effect of CXCR2 inhibitor on esophageal cell proliferation of SKP mice treated with TAM for 1 week. IHC for BrdU was performed on the section from the indicated mice i.p. injected with or without SB225002 daily in parallel to TAM administration. Scale bars, 100 μm. Bottom, quantification of BrdU+ cells. n = the number of sections from Sox2CreER/WT mice and two mice for SKPFlox/WT mice. Data represents the mean with SE. ANOVA and Dunnett’s post-hoc test were applied; *P < 0.01. (D) Soft-agar assay using human primary esophageal epithelial cells treated with recombinant IL1b and CXC ligands (25 ng/mL). Cxcls: Cxcl1, Cxcl3, Cxcl5 and Cxcl7
p53 deletion results in an invasive phenotype
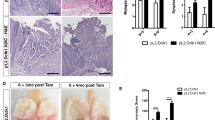
The observation that a p53 heterozygous background potentiated KrasG12D-induced hyperplastic proliferation led us to further explore the impact of homozygous p53 deletion on tumor progression. We therefore generated Sox2CreER/WT; KrasLSL-G12D/WT; p53Flox/Flox (SKPFlox/Flox) mice and treated them with TAM for 1 week. Almost all SKPFlox/Flox mice (7 of 8 TAM-treated mice) died within 2 weeks of TAM treatment. This is in contrast to SKPFlox/WT mice, which generally survived 4 weeks. SKPFlox/Flox mice that died following TAM treatment had a much larger esophagus than those of any other genotypes, including SKPFlox/WT mice (Fig. 5A and 5B). It is worth noting that invasion of GFP+ cells was only observed in the forestomach of SKPFlox/Flox mice but not SKPFlox/WT mice (Fig. 5C). A higher abundance of SASP-related factors might account for the invasive phenotype (Figs. 5D and S11), in agreement with a previous report (Coppe et al., 2008). Taken together, these results indicate that p53 homozygous deletion is required for the acquisition of an invasive phenotype.
Effect of p53 deletion on tumor progression. (A) Esophagi from different mouse models. (B and C) Lineage tracing experiment for esophagus (B) and forestomach (C) of SOX2+ cells from SKPFlox/Flox; ROSALSL-GFP/WT mice. The black arrow in Figure 5C shows invasive GFP+ tumor cells. GFP+ tumor cells, identified by a black arrow, were also observed in normal tissues on the right. Scale bars, 100 μm. (D) Effect of p53 deletion on expression of SASP-related factors. Heap map of gene expression of SASP-related chemokines described in Fig. 3E
Discussion
ASCs/PCs are found in many tissues and organs in the adult body and are important for tissue homeostasis and regeneration upon injury but, at the same time, these cells might be ideal candidates to be the cells-of-origin for cancers (Arnold et al., 2011). Here we found that SOX2+ foregut ASCs/PCs are prone to oncogenic transformation despite the presence of SOX2+ cells in other organs, such as the lungs. Our observations indicate distinct roles for oncogenic KRAS mutation and P53 deletion in tumor formation. Global gene expression analysis reveals that secreting factors contribute to the development of oncogenic KRAS-induced tumors and highlights a crucial role for the CXCR2 pathway in driving tumor formation.
SOX2 has been reported to play an important role not only in development and somatic reprogramming but also in cancer initiation/progression. For example, amplification of the SOX2 gene has been reported in human squamous cell carcinomas (SCC) of the lung and esophagus, small-cell lung cancer (SCLC) and glioblastoma (Bass et al., 2009; Annovazzi et al., 2011; Rudin et al., 2012). Overexpression of Sox2 leads to hyperplasia and tumor formation in several tissues (Lu et al., 2010; Liu et al., 2013; Mukhopadhyay et al., 2014). Furthermore, Sox2 expression marks the tumor-initiating cell population of skin squamous cell carcinomas once Sox2 expression is induced during tumorigenesis (Boumahdi et al., 2014). SOX2+ cells are also responsible for propagating medulloblastoma and targeting them prevented tumor growth (Vanner et al., 2014). Taken together, these results indicate the importance of the SOX2 molecule and SOX2+ cells in tumor development. However, tumor susceptibility of SOX2+ cells seems oncogene-specific. A previous report showed that the loss of APC in pyloric SOX2+ cells generated tumors (Sarkar et al., 2016). Similarly, targeted expression of oncogenic β-catenin in SOX2+ cells is reported to give rise to other tumor types in a non-cell-autonomous manner (e.g., pituitary tumors) (Andoniadou et al., 2013). However, we did not observe abnormal proliferation in the glandular region as well as in the pituitary in our system. These results suggest distinct oncogenic mutation susceptibilities in SOX2+ cells throughout different tissue niches.
We also found distinct roles for KRAS and P53 in oncogenic transformation of SOX2+ cells. Oncogenic Kras expression, but not p53 deletion, was sufficient to induce a hyperplasic phenotype; and p53 deletion accelerated tumorigenic proliferation in KrasG12D-induced hyperplasia. Similarly, others have found that the loss of p53 in stem cells of the colon results in tumor formation only when combined with DNA damage and chronic inflammation (Schwitalla et al., 2013; Davidson et al., 2015). Importantly, p53 homozygous deletion along with the Kras mutation led to an invasive phenotype and highly malignant tumors, highlighting the role of P53 in tumor invasion.
We identified SASP-related chemokines as responsible factors for oncogenic Kras-dependent proliferation in the forestomach and esophagus. It is thought that SASP may be induced in senescent cells to potentiate cell proliferation of surrounding pre-tumor cells and to functionally disrupt normal tissues (Krtolica et al., 2001; Coppe et al., 2008). Some of the SASP-related chemokines activate the CXCR2-dependent signaling pathway, known to trigger a secretory network that results in growth arrest, further preventing tumor progression (Acosta et al., 2008). In fact, a previous paper showed that CXCR2 is a blockade to drive oncogene-induced senescence in pancreatic tumors (Lesina et al., 2016). Inconsistent with these reports, we found that oncogenic KRAS increased the expression of SASP-related chemokines in foregut basal cells, which contributed to oncogenic proliferation. Given that epithelial cells in the esophagus and forestomach are highly proliferative, similar to pre-tumor cells, these cells might have unique characteristics, which allow them to proliferate in response to SASP-related chemokines. Interestingly, esophageal epithelial cells express some of the pluripotency factors (unpublished data), highlighting the uniqueness of these cells.
The array of genetic tumor models generated, combined with the lineage tracing experiments and global expression analyses described here, may open new paths for a better understanding of neoplasia. They may also help the future design of therapeutics targeting the initial stages of tumor formation and progression as well as facilitate the identification of novel parameters for earlier tumor diagnosis.
Materials and Methods
Mice
Mcm2CreER/WT (Pruitt et al., 2007), Sox2CreER/WT (Arnold et al., 2011), Lgr5CreER/WT (Barker et al., 2007), KrasLSL-G12D/WT (Jackson et al., 2001), p53Flox/Flox (Jonkers et al., 2001), ROSALSL-PIK3CA(H1047R)/LSL-PIK3CA(H1047R) (Adams et al., 2011), ROSALSL-Luc/LSL-Luc (Safran et al., 2003), and ROSALSL-GFP/LSL-GFP (Mao et al., 2001) have been previously described. We used both male and female mice for this study but the same gender was used for each experiment unless otherwise stated.
To activate CRE in the mice carrying CreER, TAM, dissolved in corn oil, was given orally (50 mg/mL) or intraperitoneally (20 mg/mL) to 6- to 10-week-old animals for 5 consecutive days, unless otherwise stated.
Tissue preparation and IHC
For IHC, tissues were harvested, fixed in 10% neutralized Formalin for 2 days and then stored in 70% ethanol until further processing. H&E staining, PAS staining and IHC on paraffin-sections were performed following standard protocols. The following antibodies were used for IHC: anti-GFP (Abcam, 6673, 1:200; Clontech, JL-8, 1:100); Ki67 (Cell signaling, 12202, 1:200); Proton-pump (MBL, D032-3H, 1:100); Gastrin (Santa Cruz, sc-783, 1:200); anti-p63 (Santa Cruz, sc-56188, 1:200); anti-CK13 (Abcam, 92551, 1:1000); anti-Loricrin (Abcam, 24722, 1:1000); anti-CXCL7 (Bioss Inc., A-21235, 1:200); anti-CXCR2 (Abcam, 14935, 1:200).
IVIS experiment
Mice were examined at 3 or 4 weeks post TAM administration by BLI performed using an IVIS Kinetic 2200 from Caliper Life sciences. Mice were i.p. injected with 150 mg/kg D-Luciferin (BIOSYNTH), anesthetized with isoflurane and dorsal images were then captured 10 min post luciferin injection.
RNA-sequence
Isolated tissues were homogenized with a polytron in TRIzol. The extracted RNA was purified using the RNeasy Micro Kit (Qiagen) from the homogenates. RNA quality was assessed and all samples had a minimum RNA integrity number (RIN) of 7.8. RNA library preps were prepared using the Illumina TruSeq Stranded Total RNA Sample Prep kit with Ribo-zero Gold (cat. no. RS-122-2301). Briefly, RNA was depleted of ribosomal RNA and mitochondrial RNA, then fragmented and reverse transcribed. cDNA was end-repaired, adenylated, ligated with sequencing primers and PCR amplified. Libraries were pooled and sequenced on the HiSeq 2500 using v4 sequencing reagents at single-end 50 base-pair (bp) to a depth of 15–20 million reads per experiment. Reads were mapped to the mouse genome (NCBI37/mm9) using STAR (PMID: 23104886). Gene expression levels and Gene Ontology enrichment were calculated using HOMER (PMID: 20513432) and clustering was performed using Cluster 3.0 and Java TreeView. Differential expression was defined using a false discovery rate (FDR) cut-off of 5% and a fold change of at least 2 using edgeR (PMID: 19910308). RNA-Seq data have been deposited in the Gene Expression Omnibus under accession code GSE66457.
BrdU labeling
BrdU labeling was performed using BrdU In-Situ Detection Kit (BD Biosciences, 550803) according to the manufacturer’s instructions. Briefly, the mice were i.p. injected with 1 mg of BrdU and the tissues were collected from the injected mice at 24 hr post injection, followed by paraffin embedding and sectioning. After being deparaffinized and antigen-retrieved, the section was stained using biotinylated anti-BrdU and Streptavidin HRP together with DAB substrate and BrdU+ cells were counted for quantification.
Cell culture
Mouse primary esophageal cells were derived as previously described (Kalabis et al., 2008). Briefly, the esophagi were isolated, opened longitudinally, washed in PBS followed by Dispase (1 U/mL) for 15–20 min at 37 °C. The opened esophagi were minced with forceps and incubated with TrypLE for 10 min at 37 °C. After inactivation of TrypLE with FBS, the cell suspension was filtered through 100-μm and 40-μm cell strainers. The obtained cells were centrifuged and re-suspended in keratinocyte serum-free medium (Life Technologies), followed by plating on matrigel-coated plates. Human primary esophageal epithelial cells were obtained from Cell Biologics. Het-1A cell line was obtained from ATCC. OE21 cell line was obtained from sigma. The cells were cultured according to manufacturer’s instructions.
FACS analysis
Single cell suspension of the esophagus and the forestomach was obtained as mentioned above. Lung cell isolation was performed as previously described (Gereke et al., 2012). Briefly, lungs were perfused with PBS and the salivary glands were removed to expose the trachea, followed by instillation with 1 U/mL dispase and 1% low-melting agarose. After gel solidification with ice, the lungs were isolated and washed with PBS, and incubated with dispase at room temperature for 45 min. The lungs were minced and filtered through 100-μm and 40-μm cell strainers to obtain a single cell suspension. The single cell suspension was subjected to FACS analysis.
Soft-agar assay
The cells of interest were cultured in 0.5% soft agarose layered on harder agarose in 60-mm dishes. After 14 days, the colonies were counted.
References
Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N et al (2008) Chemokine signaling via the CXCR1 receptor reinforces senescence. Cell 133:1006–1018
Adams JR, Xu K, Liu JC, Agamez NM, Loch AJ, Wong RG, Wang W, Wright KL, Lane TF, Zacksenhaus E et al (2011) Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res 71:2706–2717
Andoniadou CL, Matsushima D, Mousavy Gharavy SN, Signore M, Mackintosh AI, Schaeffer M, Gaston-Massuet C, Mollard P, Jacques TS, Le Tissier P et al (2013) Sox2(+) stem/progenitor cells in the adult mouse pituitary support organ homeostasis and have tumor-inducing potential. Cell Stem Cell 13:433–445
Annovazzi L, Mellai M, Caldera V, Valente G, Schiffer D (2011) SOX2 expression and amplification in gliomas and glioma cell lines. Cancer Genom Proteom 8:139–147
Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K (2011) Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9:317–329
Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ et al (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449:1003–1007
Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I et al (2009) SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 41:1238–1242
Blanpain C, Simons BD (2013) Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol 14:489–502
Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E et al (2014) SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511:246–250
Collado M, Serrano M (2006) The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer 6(6):472–476
Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J (2008) Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6:2853–2868
Davidson LA, Callaway ES, Kim E, Weeks BR, Fan Y-Y, Allred CD, Chapkin RS (2015) Targeted deletion of p53 in Lgr5-expressing intestinal stem cells promotes colon tumorigenesis in a preclinical model of colitis-associated cancer. Cancer Res 75(24):5392–5397
Doupe DP, Alcolea MP, Roshan A, Zhang G, Klein AM, Simons BD, Jones PH (2012) A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 337:1091–1093
Du Q, Yan W, Burton VH, Hewitt SM, Wang L, Hu N, Taylor PR, Armani MD, Mukherjee S, Emmert-Buck MR et al (2013) Validation of esophageal squamous cell carcinoma candidate genes from high-throughput transcriptomic studies. Am J Cancer Res 3:402–410
Feng Y, Bommer GT, Zhao J, Green M, Sands E, Zhai Y, Brown K, Burberry A, Cho KR, Fearon ER (2011) Mutant KRAS promotes hyperplasia and alters differentiation in the colon epithelium but does not expand the presumptive stem cell pool. Gastroenterology 141(1003–1013):e1001–1010
Gereke M, Autengruber A, Grobe L, Jeron A, Bruder D, Stegemann-Koniszewski S (2012) Flow cytometric isolation of primary murine type II alveolar epithelial cells for functional and molecular studies. J Vis Exp.
Huels DJ, Sansom OJ (2015) Stem vs non-stem cell origin of colorectal cancer. Br J Cancer 113:1–5
Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA (2001) Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15:3243–3248
Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A (2001) Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29:418–425
Kalabis J, Oyama K, Okawa T, Nakagawa H, Michaylira CZ, Stairs DB, Figueiredo JL, Mahmood U, Diehl JA, Herlyn M et al (2008) A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J Clin Invest 118:3860–3869
Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J (2001) Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A 98:12072–12077
Kuilman T, Michaloglou C, Mooi WJ, Peeper DS (2010) The essence of senescence. Genes Dev 24:2463–2479
Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS (2008) Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133:1019–1031
Lesina M, Wormann SM, Morton J, Diakopoulos KN, Korneeva O, Wimmer M, Einwachter H, Sperveslage J, Demir IE, Kehl T et al (2016) RelA regulates CXCL1/CXCR22-dependent oncogene-induced senescence in murine Kras-driven pancreatic carcinogenesis. J Clin Invest 126:2919–2932
Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding LW et al (2014) Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 46:467–473
Liu K, Jiang M, Lu Y, Chen H, Sun J, Wu S, Ku WY, Nakagawa H, Kita Y, Natsugoe S et al (2013) Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell 12:304–315
Liu QW, Fu JH, Luo KJ, Yang HX, Wang JY, Hu Y, Yang H, Bella E (2011) Identification of EGFR and KRAS mutations in Chinese patients with esophageal squamous cell carcinoma. Dis Esophagus 24:374–380
Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, Onaitis MW (2010) Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE 5:e11022
Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH (2001) Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood 97:324–326
Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A (2000) Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 14:994–1004
Mukhopadhyay A, Berrett KC, Kc U, Clair PM, Pop SM, Carr SR, Witt BL, Oliver TG (2014) Sox2 cooperates with Lkb1 loss in a mouse model of squamous cell lung cancer. Cell Rep 8:40–49
Pruitt SC, Bailey KJ, Freeland A (2007) Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells 25:3121–3132
Que J, Luo X, Schwartz RJ, Hogan BL (2009) Multiple roles for Sox2 in the developing and adult mouse trachea. Development 136:1899–1907
Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J et al (2012) Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44:1111–1116
Safran M, Kim WY, Kung AL, Horner JW, DePinho RA, Kaelin WG Jr (2003) Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging 2:297–302
Sanchez-Danes A, Hannezo E, Larsimont JC, Liagre M, Youssef KK, Simons BD, Blanpain C (2016) Defining the clonal dynamics leading to mouse skin tumour initiation. Nature 536:298–303
Sarkar A, Huebner AJ, Sulahian R, Anselmo A, Xu X, Flattery K, Desai N, Sebastian C, Yram MA, Arnold K et al (2016) Sox2 suppresses gastric tumorigenesis in mice. Cell Rep 16:1929–1941
Schwitalla S, Ziegler PK, Horst D, Becker V, Kerle I, Begus-Nahrmann Y, Lechel A, Rudolph KL, Langer R, Slotta-Huspenina J et al (2013) Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell 23(1):93–106
Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602
Shigaki H, Baba Y, Watanabe M, Miyake K, Murata A, Iwagami S, Ishimoto T, Iwatsuki M, Yoshida N, Baba H (2013) KRAS and BRAF mutations in 203 esophageal squamous cell carcinomas: pyrosequencing technology and literature review. Ann Surg Oncol 20(Suppl 3):S485–491
Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, Thompson JD, Cheng JH, Bou Reslan H, Ho CC et al (2010) Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat Biotechnol 28:585–593
Snippert HJ, Schepers AG, van Es JH, Simons BD, Clevers H (2014) Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep 15:62–69
Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y et al (2014) Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509:91–95
Tong M, Chan KW, Bao JY, Wong KY, Chen JN, Kwan PS, Tang KH, Fu L, Qin YR, Lok S et al (2012) Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res 72:6024–6035
Vanner RJ, Remke M, Gallo M, Selvadurai HJ, Coutinho F, Lee L, Kushida M, Head R, Morrissy S, Zhu X et al (2014) Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 26:33–47
Visvader JE, Lindeman GJ (2012) Cancer stem cells: current status and evolving complexities. Cell Stem Cell 10:717–728
Wang B, Hendricks DT, Wamunyokoli F, Parker MI (2006) A growth-related oncogene/CXC chemokine receptor 2 autocrine loop contributes to cellular proliferation in esophageal cancer. Cancer Res 66:3071–3077
White JR, Lee JM, Young PR, Hertzberg RP, Jurewicz AJ, Chaikin MA, Widdowson K, Foley JJ, Martin LD, Griswold DE et al (1998) Identification of a potent, selective non-peptide CXCR45 antagonist that inhibits interleukin-8-induced neutrophil migration. J Biol Chem 273:10095–10098
Zhu L, Finkelstein D, Gao C, Shi L, Wang Y, Lopez-Terrada D, Wang K, Utley S, Pounds S, Neale G et al (2016) Multi-organ Mapping of Cancer Risk. Cell 166(1132–1146):e1137
Acknowledgements
We thank Dr. Manching Ku for next-generation sequencing, Drs. Kimberly McIntyre and Varki Nissi for histology. We also thank May Schwarz and Peter Schwarz for administrative help, Marie N. Krause, Ilir Dubova, Keiichiro Suzuki, Masakazu Kurita, April Goebl, Emi Aizawa-Suzuki, Na Young Kim, and Rupa Devi Soligalla for experimental help, Elena Vicario-Orri, Toshiro Hara and Eiji Yoshihara for critical advice, and David O’Keefe and Michael Nunn for help with manuscript preparation. This work was supported by the National Key Research and Development Program of China (2015CB964800), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010100), the National Natural Science Foundation of China (81625009, 81330008, 91749202, 81861168034), Program of Beijing Municipal Science and Technology Commission (Z151100003915072), Advanced Innovation Center for Human Brain Protection (117212, 3500-1192012), and Beijing Municipal Commission of Health and Family Planning PXM2018_026283_000002). Work in the laboratory of J.C.I.B was supported by a Cancer Center Support Grant, the G. Harold and Leila Y, Mathers Charitable Foundation, The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002), The Moxie Foundation, Fundacion Dr. Pedro Guillen and Universidad Católica San Antonio de Murcia (UCAM). T.H. was supported by a Pioneer Fund Postdoctoral Scholar Award, Nomis Fellowship, and Uehara Memorial Foundation research fellowship.
Abbreviations
ASCs, adult stem cells; BLI, bioluminescence imaging; Bp, Base-pair; Cs, progenitor cells; CXCR2, CXC chemokine receptor 2; ESCC, esophageal squamous cell carcinoma; FDR, false discovery rate; HDF, human dermis skin fibroblast; hpEEC, human primary esophageal epithelial cell; IHC, immunohistochemistry; Krt17, Keratin 17; Luc, luciferase; mpEEC, mouse primary esophageal epithelial cell; PAS, periodic acid-Schiff; RNA-Seq, RNA-sequencing; RIN, RNA integrity number; SASP, senescence-associated secretory phenotype; SCC, squamous cell carcinomas; SCLC, small-cell lung cancer; TAM, tamoxifen
Compliance with ethics guidelines
Tomoaki Hishida, Eric Vazquez-Ferrer, Yuriko Hishida-Nozaki, Ignacio Sancho-Martinez, Yuta Takahashi, Fumiyuki Hatanaka, Jun Wu, Alejandro Ocampo, Pradeep Reddy, Min-Zu Wu, Laurie Gerken, Reuben J. Shaw, Concepcion Rodriguez-Esteban, Christopher Benner, Hiroshi Nakagawa, Pedro Guillen Garcia, Estrella Nuñez Delicado, Antoni Castells, Josep M. Campistol, Guang-Hui Liu and Juan Carlos Izpisua Belmonte declare that they have no conflict of interest.
All animal experiments were approved by the Committee on Animal Care at the Salk Institute. All institutional and national guidelines for the care and use of laboratory animals were followed. This article does not contain any studies with human subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hishida, T., Vazquez-Ferrer, E., Hishida-Nozaki, Y. et al. Mutations in foregut SOX2+ cells induce efficient proliferation via CXCR2 pathway. Protein Cell 10, 485–495 (2019). https://doi.org/10.1007/s13238-019-0630-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13238-019-0630-3