Abstract
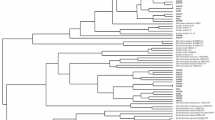
Forty-five isolates from root nodules of chickpea (Cicer arietinum L.) growing in the western and northwestern regions of Iran were characterized by phenotypic properties, repetitive element sequence-based (rep)-PCR, and 16S rRNA sequences. Based on phenotypic features, the isolates could be divided into three main groups at 92 % similarity. Nodulating isolates of those grouped in clusters II and III were grouped with reference strains of Mesorhizobium mediterraneum and M. ciceri, respectively. The three isolates of cluster I did not induce nodules on the host plant. Based on the analysis of the fingerprint patterns generated by enterobacterial repetitive intergenic consensus- and BOX-PCR, the strains were classified into six clusters (A–F) at the 75 % similarity level. These six clusters could be further divided into several subgroups at the 85 % similarity level. The phylogenetic analysis based on 16S rRNA gene sequences showed that representative isolates were closely related to M. mediterraneum, M. ciceri, and Agrobacterium tumefaciens.





Similar content being viewed by others
References
Aouani ME, Mhamdi R, Jebara M, Amarger N (2001) Characterization of rhizobia nodulating chickpea in Tunisia. Agronomie 21:577–581
Babic KH, Schauss K, Hai B, Sikora S, Redzepovic S, Radl V, Schloter M (2008) Influence of different Sinorhizobium meliloti inocula on abundance of genes involved in nitrogen transformations in the rhizosphere of alfalfa (Medicago sativa L.). Environ Microbiol 10:2922–2930
Ben Romdhane S, Aouani ME, Mhamdi R (2007a) Inefficient nodulation of chickpea (Cicer arietinum L.) in the arid and Saharan climates in Tunisia by Sinorhizobium meliloti biovar medicaginis. Ann Microbiol 57:15–20
Ben Romdhane S, Tajini F, Trabelsi M, Aouani ME, Mhamdi R (2007b) Competition for nodule formation between introduced strains of Mesorhizobium ciceri and the native populations of rhizobia nodulating chickpea (Cicer arietinum) in Tunisia. World J Microbiol Biotechnol 23:1195–1201
Ben Romdhane S, Aouani ME, Trabelsi M, De Lajudie P, Mhamdi R (2008) Selection of high nitrogen-fixing Rhizobia nodulating chickpea (Cicer arietinum) for semi-arid Tunisia. J Agron Crop Sci 194:413–420
Ben Romdhane S, Trabelsi M, Aouani ME, de Lajudie P, Mhamdi R (2009) The diversity of rhizobia nodulating chickpea (Cicer arietinum) under water deficiency as a source of more efficient inoculants. Soil Biol Biochem 41:2568–2572
Benhizia Y, Benhizia H, Benguedouar A, Muresu R, Giacomini A, Squartini A (2004) Gamma proteobacteria can nodulate legumes of the genus Hedysarum. Syst Appl Microbiol 27:462–468
Chaudhary P, Dudeja SS, Khurana AL (2001) Chickpea nodulation variants as a tool to detect the population diversity of chickpea rhizobia in soil. Physiol Mol Biol Plants 7:47–54
Chen WM, Moulin L, Bontemps C, Vandamme P, Bena G, Boivin-Masson C (2003) Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J Bacteriol 185:7266–7272
de Bruijn FJ, Rademaker J, Schneider M, Rossbach U, Louws FJ (1996) Rep-PCR Genomic fingerprinting of plant-associates bacteria and computer-assisted phylogenetic analyses. In: Stacey G, Mullin B, Greshoff P (eds) Biology of plant–microbe interaction: Proc 8th Int Congress of Molecular Plant–Microbe Interactions. American Phytopathological Society Press, St. Paul, pp 497–502
Garau G, Reeve WG, Brau L, Deiana P, Yates RJ, James D, Tiwari R, Hara GWO, Howieson JG (2005) The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant Soil 276:263–277
Ibanez F, Angelini J, Taurian T, Tonelli ML, Fabra A (2009) Endophytic occupation of peanut root nodules by opportunistic Gammaproteobacteria. Syst Appl Microbiol 32:49–55
Kantar F, Elkoca E, Ogutcu H, Algur OF (2003) Chickpea yields in relation to Rhizobium inoculation from wild chickpea at high altitudes. J Agron Crop Sci 189:291–297
Kim DH, Kaashyap M, Rathore A, Das RR, Parupalli S, Upadhyaya HD, Gopalakrishnan S, Gaur PM, Singh S, Kaur J, Yasin M, Varshney RK (2014) Phylogenetic diversity of Mesorhizobium in chickpea. J Biosci 39:513–517
Koohi A, Khodakaramian G, Zafari D (2014) Biochemical, physiological and molecular characteristics of chickpea rhizobacteria from Kurdistan province, Iran. Arch Phytopathol Plant Protect 47:919–929
Laranjo M, Machado J, Young JPW, Oliveira S (2004) High diversity of chickpea Mesorhizobium species isolated in a Portuguese agricultural region. FEMS Microbiol Ecol 48:101–107
Laranjo M, Young JPW, Oliveira S (2012) Multilocus sequence analysis reveals multiple symbiovars within Mesorhizobium species. Syst Appl Microbiol 35:359–367
Louws FJ, Fulbright DW, Stevens CT, de Bruijn FJ (1994) Specific genomic fingerprints of phytogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol 60:2286–2295
Maatallah J, Berraho EB, Munoz S, Sanjuan J, Lluch C (2002) Phenotypic and molecular characterization of chickpea rhizobia isolated from different areas of Morocco. J Appl Microbiol 93:531–540
Margaret I, Becker A, Blom J, Bonilla I, Goesmann A, Göttfert M, Lloret J, Mittard-Runte V, Rückert C, Ruiz-Sainz JE, Vinardell JM, Weidner S (2011) Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J Biotechnol 155:11–19
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214
Mhamdi R, Mrabet M, Laguerre G, Tiwari R, Aouani ME (2005) Colonization of Phaseolus vulgaris nodules by Agrobacterium-like strains. Can J Microbiol 51:105–111
Nandwani R, Dudeja SS (2009) Molecular diversity of a native mesorhizobial population of nodulating chickpea (Cicer arietinum L.) in Indian soils. J Basic Microbiol 49:463–470
Nour SM, Fernandez MP, Normand P, Cleyet-Maret JC (1994) Rhizobium ciceri sp. nov. consisting of strains that nodulate chikpea (Cicer arietinum L). Int J Syst Evol Bacteriol 44:511–522
Nour SM, Cleyet-Maret JC, Normand P, Fernandez MP (1995) Genomic heterogeneity of strains nodulating chikpea (Cicer arietinum L.) and description of Rhizobium mediterraneum sp. nov. Int J Syst Evol Bacteriol 45:640–648
Rivas R, Laranjo M, Mateos PF, Oliveira S, Martinez-Molina E, Velazquez E (2007) Strains of Mesorhizobium amorphae and Mesorhizobium tianshanense, carrying symbiotic genes of common chickpea endosymbiotic species, constitute a novel biovar (ciceri) capable of nodulating Cicer arietinum. Lett Appl Microbiol 44:412–418
Rohlf FJ (1993) NTSYS-pc: numerical taxonomy and multivariate analysis system. Version 2.0. Exeter Software, New York
Rouhrazi K, Rahimian H (2012) Genetic diversity of Iranian Agrobacterium strains from grapevine. Ann Microbiol 62:1661–1667
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sikora S, Redzepovic S, Bradic M (2002) Genomic fingerprinting of Bradyrhizobium japanicum isolates by RAPD and rep-PCR. Microbiol Res 157:213–219
Singh KB (1997) Chickpea (Cicer arietinum L.). Field Crop Res 53:161–170
Somasegaran P, Hoben HJ (1994) Handbook of rhizobia: methods in legume-rhizobium technology. Springer, New York
Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J (1996) Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev 60:407–438
Vandamme P, Goris J, Chen WM, de Vos P, Willems A (2002) Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst Appl Microbiol 25:507–512
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831
Versalovic J, Schneider M, de Bruijn FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence based PCR (rep-PCR). Methods Cell Mol Biol 5:25–40
Vincent JM (1970) A manual for the practical study of the root-nodule bacteria IBP handbook 15. Black Well Scientific Publications, Oxford
Wang LL, Wang ET, Liu J, Li Y, Chen WX (2006) Endophytic occupation of root nodules and roots of Melilotus dentatus by Agrobacterium tumefaciens. Microb Ecol 52:436–443
Weisburg WG, Barns SM, Pelletior DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wielbo J, Marek-Kozaczuk M, Mazur A, Kubik-Komar A, Skorupska A (2010) Genetic and metabolic divergence within a Rhizobium leguminosarum bv. trifolii population recovered from cover nodules. Appl Environ Microbiol 76:4593–4600
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rouhrazi, K., Khodakaramian, G. Phenotypic and genotypic diversity of root-nodulating bacteria isolated from chickpea (Cicer arietinum L.) in Iran. Ann Microbiol 65, 2219–2227 (2015). https://doi.org/10.1007/s13213-015-1062-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-015-1062-9