Abstract
A unique symbiosis occurs between embryos of the spotted salamander (Ambystoma maculatum) and a green alga (Oophila amblystomatis). Unlike most vertebrate host-symbiont relationships, which are ectosymbiotic, A. maculatum exhibits both an ecto- and an endo-symbiosis, where some of the green algal cells living inside egg capsules enter embryonic tissues as well as individual salamander cells. Past research has consistently categorized this symbiosis as a mutualism, making this the first example of a “beneficial” microbe entering vertebrate cells. Another closely related species of salamander, Ambystoma gracile, also harbors beneficial Oophila algae in its egg capsules. However, our sampling within the A. gracile range consistently shows this to be a strict ectosymbiotic interaction—with no sign of tissue or presumably cellular entry. In this study we swapped cultured algae derived from intracapsular fluid of different salamander hosts to test the fidelity of tissue entry in these symbioses. Both A. maculatum and A. gracile embryos were raised in cultures with their own algae or algae cultured from the other host. Under these in vitro culture conditions A. maculatum algae will enter embryonic A. maculatum tissues. Additionally, although at a much lower frequency, A. gracile derived algae will also enter A. maculatum host tissues. However, neither Oophila strain enters A. gracile hosts in these co-culture conditions. These data reveal a potential host-symbiont fidelity that allows the unique endosymbiosis to occur in A. maculatum, but not in A. gracile. However, preliminary trials in our study found that persistent endogenous A. maculatum algae, as opposed to the cultured algae used in subsequent trials, enters host tissues at a higher frequency. An analysis of previously published Oophila transcriptomes revealed dramatic differences in gene expression between cultured and intracapsular Oophila. These include a suite of genes in protein and cell wall synthesis, photosynthesis, central carbon metabolism suggesting the intracapsular algae are assimilating ammonia for nitrogen metabolism and may be undergoing a life-cycle transition. Further refinements of these co-culture conditions could help determine physiological differences between cultured and endogenous algae, as well as rate-limiting cues provided for the alga by the salamander.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Past experimental studies have established the Oophila amblystomatis—Ambystoma maculatum symbiosis as a mutualistm (Gilbert 1942, 1944; Bachmann et al. 1985; Mills and Barnhart 1999; Pinder and Friet 1994; Bianchini et al. 2012). Recently, this association was revealed to also include tissue and even cellular entry by Oophila during A. maculatum development (Kerney et al. 2011; Burns et al. 2017). Algae proliferate outside the blastopore during Harrison (1969) Stages 15–17 and are first found entering host tissues by Stage 26. To date, this tissue and cellular entry of a beneficial algae is unique to A. maculatum among the vertebrates.
1.1 Diversity of “Oophila”
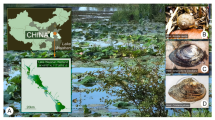
Oophila clade algae also associate with at least four species of North American amphibians including two species of Rana frogs and two species of Ambystoma salamanders (Kerney 2011; Kim et al. 2014). More recently Oophila amblystomatis from a unique independent subclade have been described from the egg capsules of the Japanese salamander Hynobius nigrescens (Muto et al. 2017). Studies have found that the Oophila alga living in association with the closely related Ambystoma gracile is also beneficial to its host (Marco and Blaustein 2000). However, unlike A. maculatum, preliminary analyses have failed to find similar algal entry into host tissues of A. gracile (Kerney et al. 2017; Fig. 1).
Both Ambystoma maculatum (a–d) and Ambystoma gracile (e–h) have intracapsular Oophila amblystomatis algae living in association with their embryos (c, g) despite their geographic isolation (b, f). While this alga can enter tissues and cells of A. maculatum (d white, arrowheads), it has never been observed embedded in A. gracile tissues (h, N = 10 clutches). Range maps in (b) and (f) Copyright © 2018 NatureServe (IUCN et al. 2004).
Together the Japanese and North American Oophila form a monophyletic group within the Moewusii clade of Chlamydomonodales that includes three species of free-living algae (Kim et al. 2014; Muto et al. 2017). There are five subclades within the larger Oophila clade (Kim et al. 2014; Muto et al. 2017). Algae from well-resolved subclades I and II have only been found to associate with Ambystoma maculatum and Ambystoma gracile. To date we have only found algae from subclade I within Ambystoma macualtum embryonic tissues and cells (Kerney et al. 2011; Burns et al. 2017). Algae from the subclades III and IV (clade B of Nema et al. 2018) have been found living in association with A. maculatum, the North American frogs Rana (Lithobates sic) sylvatica and R. aurora (Kim et al. 2014), as well as A. gracile (this study). The Japanese subclade J1 and the free-living algae Chlamydomonas nasuta cluster with subclade III (Muto et al. 2017).
Recently Nema et al. (2018) have suggested a polyphyly of the Oophila clade based on a ribosomal SSU from A. maculatum egg capsules collected from five sites in Ontario, Canada, a sub-sampling of published Oophila sequences, and unpublished sequences of Oophila that were collected from multiple sites in New England (Lewis and Landberg 2014). Oophila algae from subclades I through IV and Japanese subclade J1 (Kim et al. 2014; Muto et al. 2017) would all likely fall into “Clade” B of Nema et al. 2018 (although J1 sequences were excluded from their analysis). In co-culture experiments, we used algae from subclades I (associated with A. maculatum) and II (associated with A. gracile) (Kim et al. 2014; Muto et al. 2017), which we designate as “Oophila” here (following Schultz 2016). Prior to this study it was unknown whether or how the algal cells outside subclade I would interact with A. maculatum in controlled laboratory co-cultures.
1.2 Co-cultures
The manipulation of host-microbe co-cultures is a powerful tool in symbiosis research. By recombining populations of microbial cells with endogenous or novel hosts, it is possible to test host and microbial specificity in mutualist associations. This specificity can have implications for mechanisms of symbiont acquisition and maintenance as well as indicate potential co-evolutionary dynamics between the two partners.
In other systems, co-culture approaches have been employed to reveal the specificity of the facultative ectosymbiotic Euprymna—Allivibrio associations (Mandel et al. 2009), facultative endosymbiotic Hamiltonella defensa bacteria in aphids (Oliver et al. 2010), and facultative endosymbiotic dinoflagellate mutualists in coral (Voolstra et al. 2009). The approach has also been used to investigate obligate associations. Advances in culturing and experimental methodologies have allowed the transfer of obligate endosymbiotic Buchnera strains into new aphid matrilines (Moran and Yun 2015), and the replacement of the endogenous eukaryotic alga with a novel cyanobacterial partner in Paramecium bursaria (Ohkawa et al. 2011). Similar approaches can even establish symbionts in entirely novel hosts, such as the permanent lines of vertically inherited Wolbachia strains in novel Aedes mosquito hosts (Walker et al. 2011). These host-symbiont exchanges often help determine the extent to which these co-evolved physiological relationships are integrated between symbiont and host.
Co-culture experiments require established culture conditions for both partners in a symbiotic association. In the Ambystoma—Oophila association, both host and symbiont can be cultured in isolation. Pioneering embryologists developed protocols for Ambystoma maculatum embryo rearing (then Amblystoma punctatum) for embryonic tissue ablation and transplant experiments (Burr 1916; Twitty 1932), while established culturing protocols for Oophila sp. (Cliburn and Ward 1963) have recently been refined for taxonomy and cultured toxicology studies (Kim et al. 2014; Rodriguez-Gil et al. 2014).
Earlier work has shown that algae proliferate outside the blastopore during early neurulation (Harrison stage 15). Algae enter the embryonic tissues after this proliferation, between Stages 17 and 26, and are detectable from algal autofluorescence into the early larval stages in A. maculatum (Kerney et al. 2011). Similar algal entry was not detected in stage-matched A. gracile (Kerney et al. 2017), however this was only a preliminary observation.
The current study aims to determine 1) the extent to which algae enter field-collected A. gracile hosts, 2) whether Oophila derived from an A. gracile host will enter A. maculatum hosts in experimental co-cultures, 3) whether Oophila derived from A. maculatum host will enter A. gracile hosts in experimental co-cultures and 4) the extent to which algae from different subclades benefit from being co-cultured with their endogenous host in comparison to a novel partner. During our experiments we found that persistent native algae that were not sufficiently removed from early host embryos could enter tissues at a higher rate than the cultured algae used in this experiment. Therefore we included the additional aim of comparing the transcriptional profile of native intracapsular algae with algae raised in lab cultures of AF6 media. These aims were achieved through a combination of field collections, algal culturing, analysis of published RNA sequence libraries (Burns et al. 2017), and controlled co-cultures of field-collected salamander embryos at early developmental stages.
2 Methods
2.1 Algal culturing
Algal culturing techniques are described in Kim et al. (2014). All algae were maintained in AF6 media (Wantanabe et al. 2000) in a Conviron Environmental chamber set at 18 °C on a 12 h light/ 12 h dark timer with 59 μmol/m2/s (PAR) broad-spectrum fluorescent light. We also experimented with the NH4+ supplemented Bristol’s media described in (Rodriguez-Gil et al. 2014) however in our experience Oophila grew better in AF6. Importantly, these algal lines were established without plating the cultures on agar solidified media. As explained previously, this process enriches for non-Oophila algae as Oophila does not grow well on agar plates (Kim et al. 2014). Use of solidified media may be biasing the reported polyphyly of Oophila proposed in a conference proceeding (Lewis et al. 2013). The strain of A. maculatum derived alga used in this study is from Halifax Nova Scotia and belongs to subclade I. Based on 18S rDNA sequence analysis, the A. gracile derived algal strain used in this study is from Washington State and belongs to subclade III (following Kim et al. 2014). All algal strains were sub-cultured at a 1:100 dilution into fresh AF6 for between two to three weeks prior to the experimental trials.
2.2 Field collections
None of the amphibian embryos used in these experiments are endangered or specially protected by state or federal laws. Collections were approved as part of an Institutional Animal Care Protocol to R Kerney (IACUC#2013F17; Gettysburg College Animal Care Committee). All collections were made under state-specific permits as indicated in Table S1. Ten clutches of A. gracile embryos were collected across ten sites in California, Oregon, and Washington (see Table S1).
2.3 Screening Ambystoma gracile for algal entry
Ten field-collected clutches with embryos between Stages 25–36 were screened for algal autofluorescence in the field with a Nikon Eclipse TS100 fluorescent microscope fitted with a custom chlorophyll filter cube from Chroma Technology Corporation (excitation 480/40 nm; emission 600 nm long pass) and a 10X objective lens immediately after collection. Representatives of 1–2 embryos per clutch were returned to the lab for vibratome sectioning (300 μm increments, 1000Plus Pelco 102 Vibratome, Ted Pella Inc., Redding, CA) and subsequent screening following the co-culture microscopy protocols described below.
2.4 Co-culture sampling
Live embryos from 3 of the ten A. gracile clutches that were screened for algal entry in the field were shipped to Gettysburg College for reciprocal co-culturing experiments with previously-cultured Oophila algae raised in AF6 media from A. gracile or A. maculatum hosts. The clutches were from three separate sites (Table S1), and each contained embryos in early developmental stages (Harrison Stages 7–12).
2.5 Co-culturing
Wild-collected embryos were transferred to sterilized 20% Holtfreter’s solution (Sive et al. 2000) and removed from their egg capsules with sterilized watchmaker forceps. The embryos were then treated with 0.01% formalin to minimize growth of either endogenous eukaryotic algae or prokaryotes on the surface of the embryo, followed by six one-minute rinses in fresh sterilized 20% Holtfreter’s solution (modified from Gibbs 2003). These rinses and subsequent co-cultures were carried out in sterile 24-well plates with 2 ml liquid volume.
Embryos were co-cultured with algal cells in 2mls of 10% Holtfreter’s and 50% AF6. Earlier trials in 100% AF6 led to complete mortality of the salamander embryos, potentially due to the high levels of nitrates (1.9 mM) and ammonia (275 μM) in the solution (data not shown). Field-collected salamander embryos ranged from Stages 7–12 at the beginning of the co-cultures. These stages are prior to the detectable algal bloom in field-collected embryos (stage 15, Kerney et al. 2011) and prior to detectable chlorophyll (based on spectrophotometer readings of acetone extracts) inside the egg capsule fluid (Stage 18, Small et al. 2014).
The initial algal cell concentrations varied from 200 to 500 algal cells/μl at the beginning of the co-culture trials. This variation was due to the slow growth of algal cultures and tight time constraints of these seasonal experiments. Preliminary data has revealed low recovery (maximally 26%) under moderate centrifugal force and by high mortality with greater centrifugal force from concentrating algae with centrifugation (J. Burns personal observation). Since all co-cultures were established with relatively high numbers of exogenous algae prior to detectable endogenous chlorophyll (Small et al. 2014) we were not concerned with this potentially confounding variable. Subsequent analyses revealed that the extent of algal proliferation did not correlate with final algal entry (see binary logistic regression results below).
Embryos were incubated from 10 to 14 days in a Conviron Environmental chamber set at 15 °C on a 12 h light/ 12 h dark timer with 59 μmol/m2/s broad spectrum fluorescent light. Salamanders ranged from Harrison Stages 34–39 at the end of the co-culture experiments. This developmental interval spans the bloom of algal cells observed outside the blastopore (Stage 15) and initial detection of tissue entry by algal cells (Stage 26) and peak detectable intra-tissue algae observed in Ambystoma maculatum (Kerney et al. 2011; Small et al. 2014).
Specimens were fixed overnight in 10% neutral buffered formalin and stored in phosphate buffered saline (PBS) at 4 °C. These were assayed with a fluorescent microscope (Nikon Eclipse 90i) for algal entry into tissues within three weeks post fixation (Y-2E/C, Texas Red bandpass filter 540-580 nm excitation 600-660 nm emission, 10X objective). All embryos were manually bisected along the sagittal midline with a razor to reveal deep autofluorescence or potential algae in the alimentary canal. Whole mount specimens with detectable auto-fluorescence were embedded in 1% agarose in PBS, vibratome sectioned (300 μm), and individual sections were imaged again with 6 sec exposures. Monochrome fluorescent images were normalized for brightness and contrast using the “auto level” function in Adobe Photoshop (CS5) to standardize image contrasts prior to scoring detectable autofluorescence in sectioned tissues.
Each clutch was divided into three group of embryos. These were co-cultured with 1) A. gracile derived algae, 2) A. maculatum derived algae, or 3) no-algae negative controls. Several trials in 2015 were contaminated with endogenous algae due to shorter, 5 min, formalin treatments initially used in those experiments (see below). Tissue entry data from these shorter formalin treatment trials were not used in our co-culture results although these negative controls were screened for the extent of endogenous algal cell entry into host tissues.
2.6 Statistics
All statistics were performed in R for Mac OSX (V. 3.3.3). Influence of algal growth, host type, and algal strain on tissue entry was compared with a binomial logistic regression using the GLM command. Tissue entry was treated as a binary dependent variable. Pairwise differences in final algal concentrations and algal proliferation was analyzed using two-tailed, type-II, Student’s T-tests. Pairwise comparisons of survival and frequency of tissue entry employed Pearson’s Chi squared.
2.7 Differential expression analysis
Preliminary trials with shorter formalin treatments (5 min) found that endogenous A. maculatum algae enters host tissues in a higher percentage of trials than cultured algae. We analyzed a published transcriptomics dataset (Burns et al. 2017) to explore physiological differences between cultured and intracapsular A. maculatum derived algae (subclade I Kim et al. 2014). Transcriptome assembly and differentially expressed transcripts with associated annotations were mined from data collected for Burns et al. (2017). For the current manuscript, we completed a comparison between mRNA expression levels in cultured O. amblystomatis (UTEX LB3005, n = 4 cultures) and intracapsular algae extracted from egg capsules collected in Gettysburg, PA (n = 3 egg capsules from Harrison stage 38 embryos). Transcript GC content and abundance normalization was conducted as in Burns et al. 2017. Differential expression analysis was conducted in edgeR (V. 3.22.3) (Robinson et al. 2010). Gene ontology (GO) analysis was conducted in topGO (V. 2.32.0) (Alexa and Rahnenfuhrer 2016) using a custom gene association file for O. amblystomatis transcripts annotated against UniProt/SwissProt as in Burns et al. 2017. Significantly enriched GO categories were visualized using the GOplot package (V. 1.0.2) (Walter et al. 2015).
2.8 PAGE gel of intracapsular fluid
Intracapsular fluid was extracted by aspiration with a syringe and 23-gauge needle from eggs containing Harrison stage 38 embryos. The extracted fluid containing intracapuslar algae and other less abundant microbes and debris was centrifuged at 14,000 x g for 10 min. The supernatant was recovered and centrifuged again at 14,000 x g for 10 min. The supernatant was recovered and mixed directly with Laemmli loading buffer (Laemmli 1970) and loaded onto an 8% denaturing PAGE gel. Protein bands were visualized with Coomasie blue dye.
3 Results
3.1 Ambystoma gracile embryos
None of the 270 Ambystoma gracile embryos observed from individual clutches collected from seven sites had detectable algal autofluorescence inside host embryonic tissues between Stages 25–36 when screened by whole embryo fluorescence microscopy. Additionally, of the 35 A. gracile embryos manually dissected with a scalpel or vibratome-sectioned only a single Stage 36 specimen collected from Yachats, OR had detectable autofluorescence inside the alimentary canal. However, no algal cells were embedded inside any of the embryonic host tissues (Fig. 1h).
3.1.1 Ambystoma gracile embryos with A. maculatum derived Oophila
A. gracile embryos were co-cultured with 500 algal cells/μl of A. maculatum derived algae beginning at Stage 12 and ending at Stages 35–37 over a 14 day period. Of the original 48 embryos, 35 survived to the end of the experiment (73%). None of these exhibited algal autofluorescence inside host tissues in vibratome sections (Table 1, Fig. 2).
Representative results of reciprocal co-cultures. Tissue entry was only observed in A. maculatum embryos (a), with a higher percentage of A. maculatum derived algae entering host tissues. Only one A gracile embryo (b) exhibited algal entry into the alimentary canal in our wild-caught samples however there was no sign of tissue entry from this cavity. None of the A. gracile co-cultures exhibited either alimentary or tissue entry (c)
3.1.2 Ambystoma gracile embryos with A. gracile derived Oophila
Ambystoma gracile embryos were co-cultured with 500 algal cells/μl of A. gracile derived algae beginning at Stage 12 and ending at Stages 30–36 over a 14 day period. Of the original 48 embryos, 22 survived to the end of the experiment (46%). None of these exhibited algal autofluorescence inside host tissues in vibratome sections (Fig. 2).
3.1.3 Ambystoma gracile negative controls
There was high mortality in the A. gracile negative controls. Of the 18 initial Stage 12 embryos only three survived to the end of the 14-day trial (17%). These three were between Stages 31–32 and none contained any residual algae.
3.2 Ambystoma maculatum embryos
3.2.1 Ambystoma maculatum embryos with A. maculatum derived Oophila
Ambystoma maculatum embryos were co-cultured with 200 algal cells/μl of A.maculatum-derived algae beginning at Stages 7–9 and ending at Stages 35–37 over a 12-day period. Of the original 111 embryos, 39 survived to the end of the experiment (35%). Fifteen of these exhibited algal autofluorescence inside host tissues in vibratome sections (39% of surviving) under these experimental conditions. Algal autofluorescence was distributed in derivatives of all three primary germ layers including the somites, lateral plate mesoderm, notochord, yolk, neural tube, head mesenchyme, and dermis. The highest amount of algae was concentrated in the yolky endoderm and algae were found in the alimentary canal of nine out of the fifteen embryos that contained algae in their tissues. Unlike A. gracile, no A. maculatum embryos had algae restricted to their alimentary canal but excluded from their tissues.
3.2.2 Ambystoma maculatum embryos with A. gracile derived Oophila
Ambystoma maculatum embryos were co-cultured with 320 algal cells/μl of A. gracile derived algae beginning at Stage 12 and ending at Stages 36–37 over a 10-day period. Of the original 134 embryos, 113 survived to the end of the experiment (84%). Two of these exhibited algal autofluorescence inside host tissues in vibratome sections (1.6%) under these experimental conditions. One had scattered algal cells in head mesenchyme, the neural tube, and dermis. The other had diffuse algal cells in the yolk, somites, notochord and neural tube. Two additional embryos exhibited algal autofluorescence inside the host alimentary canal, with no sign of algal cell entry into host tissues.
3.2.3 Ambystoma maculatum negative controls
Of the 72 negative control A. maculatum from successful trials (with complete removal of endogenous algae), 39 survived to the end of the experiment (54%). None of these exhibited algal growth inside the experimental wells or algal entry into host tissues.
3.2.4 Algal growth in response to different hosts
Almost all cells counted at the end of the co-cultures were putative zoospores, none were the larger putative zygotes, regardless of algal type, host, or tissue entry (following Bishop and Miller 2014). Only one experimental condition, A. gracile with A. maculatum algae, had a detectable amount of zygotes in the final algal culture (0.19% of the total algal count) based on haemocytometer counts.
There was a significant difference in algal proliferation between the two algal strains (end concentration/beginning concentration), regardless of the host used in the co-culture experiment. A. maculatum algae proliferated to a greater extent (4.80 fold increase, SEM: 0.78) than the A. gracile derived algae (1.57 fold increase, SEM: 12.76) in the presence of either host (2-tailed t-test, df = 196, t-calc = −9.0, P < 0.005). However, there was no difference in the fold change proliferation of algae between the hosts (ANOVA, df = 1, F = 0.26, P = 0.61).
3.2.5 Variables accounting for tissue entry
A binary logistic regression was used to test the influence of algal strain (A. gracile or A. maculatum), clutch survivorship (percent survival) and algal proliferation (fold change) on the dependent binary variable of tissue entry within the A. maculatum trials (N = 152 embryos screened for tissue entry). None of the variables had a significant influence on the probability of tissue entry within this model. However, a chi-square based analysis of deviance showed that algal strain (A. maculatum or A. gracile derived) significantly reduced the residual deviance (Χ2 = 192, df = 136, P < 0.001) in comparison to the null deviance model (a model which only shares the intercept). This indicates that algal strain alone significantly improves the model’s prediction of tissue entry. Neither clutch survivorship, algal proliferation, nor any interacting variables improved the model (Χ2 = 125, df = 151, P > 0.5).
3.2.6 Negative controls retaining endogenous algae in early trials
Initial trials of A. maculatum and A. gracile co-cultures had to be discarded as there was insufficient sterilization of endogenous algae in the negative controls. This was due to a five-minute soak in 0.01% formalin as opposed to the longer ten-minute soak that was used in subsequent trials (Fig. 3).
The A. maculatum negative control trial with persisting algae began with 24 embryos, of which 19 survived to Stages 29–32 (79%). The final algal concentration increased to a range of 10–220 cells/μl. Of these 3.4% were zygotes. Fourteen of the surviving embryos exhibited tissue entry of algal cells (74%). In comparison to the A. maculatum derived algal co-cultures the survival (Pearson’s chi-square test, df = 1, P < 0.001) and extent of algal cell entry (Pearson’s chi-square test, df = 1, P < 0.001) were both significantly higher.
The A. gracile negative control trial with persisting algae began with 24 embryos. Of these, 17 survived to Stages 35–37 (71%). A. gracile algal concentrations increased from being undetected to a range of 50–1000 cells/μl. None of these algae were non-motile zygotes (all had flagella), and none of the embryos exhibited any sign of tissue or alimentary canal entry. In comparison to the A. gracile derived algal co-cultures the embryo survival was significantly higher (Pearson’s chi-square test, df = 1, P = 0.01).
3.2.7 Differential expression analysis
Given the observed differences in tissue entry of endogenous algae compared to algae grown in lab culture, we examined transcriptional differences between intracapsular algae and cultured algae. This analysis is based on data generated in Burns et al. (2017). Intracapsular algae transcriptomes are from stage 38 A. maculatum embryos, during peak tissue invasion. We see 2937 over-expressed and 1422 under-expressed transcripts in intracapsular algae relative to algae grown in AF6 lab culture (Fig. 4). Of those, 731 (25%) of the over-expressed and 672 (47%) of the under-expressed transcripts contained open reading frames that could be annotated against the Uniprot-SwissProt database.
Dotplot of relative mRNA expression between intracapsular and cultured alga. Each dot represents one O. amblystomatis transcript. Abbreviations: logFC, base 2 logarithm of fold change between intracapsular and cultured algae; logCPM, base 2 logarithm of individual transcript counts per million sequencing reads. Blue lines indicate transcripts with a 2-fold change up or down in intracapsular algae relative to cultured algae. Red dots indicate transcripts with false discovery rate adjusted p < 0.05 (negative binomial regression, Benjamini and Hochberg FDR ).
3.2.8 Gene ontology (GO) analysis reveals changes to core algal machinery
GO term analysis was used to summarize functional processes represented among differentially expressed genes. Differentially expressed genes are enriched in core cellular processes like protein synthesis (translation), photosynthesis, and central carbon metabolism (Fig. 5). Notably, Krebs/ cirtic acid/ tricrboxylic acid (TCA) cycle genes were largely downregulated, while genes involved in cellulose biosynthesis and translation were largely upregulated (Fig. 5).
GO term analysis reveals changes to central metabolic processes. Plot of the most significantly enriched GO terms and the relative expression of significantly over- and under-expressed genes for each category. The Z-score is a measure of the proportion of over- and under-expressed genes in a category
4 Discussion
Our results show a degree of symbiont and host specificity for the tissue entry of Oophila amblystomatis into Ambystoma maculatum embryos. Of the two species assayed, only A. maculatum acquires algal cells during its embryonic development. Only a single field-collected A. gracile exhibited alimentary canal entry of algal cells, and none of the wild or experimental A. gracile exhibited tissue entry of either Oophila strain. Additionally, the endogenous algae cultured from an A. maculatum host entered tissues at a higher frequency (38.5%) than algae derived from an A. gracile host (1.6%). We screened far more A. maculatum embryos with algae cultured from A. gracile hosts (117 co-cultures), than A. maculatum embryos raised with algae derived from A. maculatum hosts (39 co-cultures), to find only two embryos with A. gracile algae inside their tissues. These data are consistent with either an active host recruitment of algal cells into A. maculatum, or a tolerance for tissue-invading algae by A. maculatum. Neither of these potential features apply to A. gracile hosts with algae derived from A. gracile clutches (22 co-cultures) or with algae derived from A. maculatum clutches (35 co-cultures) as no tissue entry was seen in either case. In addition, 270 A. gracile embryos were screened by whole embryo long exposure fluorescence microscopy, and none had evidence of algal fluorescence in host tissues. This is in contrast to previously reported screening of A. maculatum embryos with 86.6% containing detectable algae in host tissues by stage 38.8 (± 0.1) using the same sectioning and screening methods (Small et al. 2014).
4.1 Oophila life history inside the egg capsule
Oophila amblystomatis has a dynamic life history inside the embryonic egg capsules. The extent to which symbiotic Oophila are acquired from the environment is still unclear. Recently free-living Oophila have been detected within vernal pools (Lin and Bishop 2015) although the pathway of their selective recruitment into egg capsules is unknown. These algal cells appear as either bi- or quadri-flagellated zoospores (presumably post-syngamy) or larger circular putative zygotes (Goff and Stein 1976; Bishop and Miller 2014; Kim et al. 2014). Early putative zygotes have been observed on the innermost egg capsule soon after laying (Kerney et al. 2011). These are surrounded by an outer envelope that is similar to those surrounding encysting diatoms (Dodge 1973). The chlorophyll concentration of the egg capsule fluid increases from Stages 12 through 40 before falling off by Stage 43 (Small et al. 2014 ). This increase coincides with a dramatic proliferation of motile zoospores around the embryonic blastopore by Stages 15–17 (Kerney et al. 2011; Bishop and Miller 2014). Following this proliferation, some of the algae either enter embryonic tissues (Kerney et al. 2011) or undergo syngamy to become putative zygotes, which aggregate further on the egg capsule wall (Bishop and Miller 2014). Surprisingly, these putative zygotes are not behaving like the photosynthetically dormant zygospores of Chlamydomonas rheinhardti, but instead remain photosynthetically active while bound to the egg capsule (Baldan et al. 1991; Bishop and Miller 2014). Approximately 9.6% of the egg capsule algae are these large putative zygotes by Stage 37 (based on regressions from Bishop and Miller 2014). Meanwhile the number of zoospores in the intracapsular fluid remains relatively constant through the embryonic period but drops off closer to hatching (Stage 40; Bishop and Miller 2014).
Our experimental set up had some interesting discrepancies with the progression the Oophila- A. maculatum relationship as it occurs inside the egg capsules. The number of algal cells found in the experimental wells varied, with a higher prevalence and growth rate of A. maculatum derived algae. In field-collected clutches, the Stage 36 capsule volume is approximately 200 μl (Small 2014 their Fig. 2). The number of Stage 36 algal cells is 189,470 cells per egg capsule (Bishop and Miller 2014, their Fig. 2a) giving an approximate algal concentration of 947 cells/μl. This is less than the final concentration of A. maculatum derived algae in our co-culture conditions (appx. 2000 cells/μl for both hosts). However, it is higher than the final concentrations of co-cultured A. gracile derived algae (435 cells/μl with A. maculatum and 834 cells/μl with A. gracile). While the A. maculatum derived alga was also more likely to enter tissues, our logistical regression analysis did not find algal proliferation as an explanatory variable for tissue entry. This is supported by the two A. gracile derived algae trials that entered A. maculatum hosts. Both trials had lower algal proliferation (1.1 fold and 0.9 fold respectively) in comparison to the average A. gracile derived algae proliferation with A. maculatum hosts (1.6 fold).
Our final in vitro algal populations were overwhelmingly composed of flagellated zoospore algae with almost no detectable zygotes in any of our final samples. The two exceptions were a small number of zygotes surrounding A. gracile embryos with A. maculatum algae (0.19% putative zygotes) and the negative control trial with A. maculatum embryos where endogenous algae persisted following our initially shorter pre-trial “sterilization.” These comprised 3.4% zygotes in the final algal populations associated with the embryos (Stages 35–37). While this is less than the 9.6% reported by Bishop and Miller (2014) this is still a dramatic increase in the number of zygotes over the cultured algae experiments.
4.2 Host mortality
Our study encountered much higher host mortality than recent research on isolated rearing of Ambystomatid embryos (Small et al. 2014; Hale et al. 2016, 2017). This discrepancy is likely attributable to our process of removing endogenous algae in a 0.01% formalin solution. This removal was critical for our experimental co-cultures. Our protocol is modified from an axolotol (Ambystoma mexicanum) protocol for testing drug exposures (Gibbs 2003), and includes the longer 10-min soak compared to a less stressful 5-min soak in 0.01% formalin. The shorter 5-min soak resulted in persistent endogenous algae (dubbed our “failed negative control”) which would have confounded our co-culture results as endogenous algae and exogenously added cultured algae are visually indistinguishable. A 5-min soak in 0.01% formalin also resulted in significantly lower embryonic mortality in both A. maculatum and A. gracile. Given the apparent stress caused by the longer 10-mintute formalin exposure we deliberately excluded any potential host-benefit analysis from this project.
4.3 Algal entry into hosts
The rates of A. maculatum algal cell entry into their endogenous hosts are comparable to those reported in the literature for wild collected clutches. Small et al. (2014) found between of 27–87% of embryos assayed containing algae inside their tissues between Stages 30 and 40 (50% average), as detected by fluorescence microscopy on 300 μm vibratome sectioned samples. Using similar detection techniques we found 39% with tissue entry in our experimental co-culture conditions. However, this increased to 74% in our failed negative control.
The failed negative control trials indicated that endogenous algal cells are physiologically different from the AF6 cultured algae used in our experiments. This is also in agreement with the general lack of circular zygotes in our final co-culture conditions. These algae persisted from early developmental stages and managed to populate the 1 ml AF6/Holtfreter’s solution used in our trials. This further suggests that the physiological difference between endogenous capsular algae and AF6 reared cultures is established by early developmental stages, prior to the establishment of our co-cultures. This is also prior to algal proliferation around the blastopore.
4.4 Algal gene expression in the intracapsular environment
Using transcriptome data from Burns et al. (2017) we analyzed the differential expression of A. maculatum derived algal transcripts from intracapsular and AF6-cultured algae. Gene ontology (GO) grouping of the differentially expressed genes revealed some intriguing differences in the transcriptional profile between intracapsular algae and cultured algae (Fig. 5). Strikingly, intracapsular algae have relative under-expression of central carbon metabolism genes in glycolysis and the TCA cycle. One notable pattern in central carbon metabolism is upregulation of phosphoenolpyruvate carboxylase 1 (PEPC) gene and downregulation of the first subunit of pyruvate dehydrogenase (PDH) (Table 2). These changes are indicative of altered flux through the TCA and an increase in anaplerotic (replenishing TCA intermediates) reactions where pyruvate is converted directly to the TCA intermediate oxaloacetate (Owen et al. 2002). In plants and green algae, increased flux through PEPC is associated with ammonia assimilation (Guy et al. 1989), and in this symbiosis, ammonia is thought to be the nitrogen source for intracapsular algae (Small et al. 2014). Other ammonia assimilation genes, such as glutamate synthetase (GOGAT) and a 2-oxoglutarate/malate transporter (DIT1) were also relatively overexpressed in intracapsular algae. Additional ammonia assimilation genes such as glutamine synthetase (GS) and glutamate dehydrogenase (GDHA) and a slew of TCA enzymes, however, were relatively under-expressed in intracapsular algae (Table 2) indicating a need for further investigation into this pathway.
The intracapsular algae display enrichment of overexpressed genes involved in protein synthesis and cellulose biosynthesis (Fig. 5). As the intracapsular algal samples were collected from eggs containing stage 38 embryos, around peak algal density, these changes might reflect a life stage transition for the algae where nitrogen is becoming limiting and the cell wall is thickening in response (Small et al. 2014). A subset of Oophila in A. maculatum egg capsules undergo syngamy beginning at stage 25 and adhere to the egg capsule wall through post-hatching (Bishop and Miller 2014). Some of the transcriptional changes we are observing in this analysis may be attributable to this life history transformation occurring in our intracapsular sample, while the majority of cultured algae remain flagellated zoospores.
There are four upregulated transcripts in intracapsular algae that are annotated as gamma-glutamyl hydrolases (GGH) (Table 2) that are typically vacuolar enzymes in plants used to break down folylpolyglutamates (Orsomando et al. 2005; Akhtar et al. 2010; Ravanel et al. 2011). In plant tissues, upregulation of GGH leads to a release of folate from plant vacuoles and lowers their overall folate content (Jeong et al. 2017). The intracapsular algae may be similarly mobilizing vacuolar folate stores. Overexpression of genes involved in regulating redox potential, such as antioxidant proteins in the glutaredoxin and thioredoxin groups (Table 2), may relate to the hyperoxic environment created by the intracapsular algae during daylight hours (Pinder and Friet 1994).
Notably, the intracapsular fluid that contains both the developing embryo and its symbiotic algae also contains many extracellular proteins detectable with Coomassie staining (Fig. S1). These algae may be using proteins or possibly small organic molecules in the intracapsular fluid to support a form of mixotrophic growth, influencing the observed changes to central carbon metabolism.
A potential confounder for the differential expression analysis is that the native intracapsular algae may include multiple Oophila strains, while the cultured algae is clonal (subclade 1). This could lead to different baseline transcript levels across many genes. Indeed, it was found that more than one Oophila strain can co-exist within the same egg capsule (Kim et al. 2014). We might, however, expect natural strain differences to result in fewer differentially expressed genes than observed here (Carrier et al. 2014). Moreover, the only 18S rRNA fragments assembled in the intracapsular algae transcriptomes are from Oophila clade I, the same clade as the cultured alga used for comparison. Finally, the statistical support for the differential expression calls required consistency in read counts of each transcript pool across three biological replicates of intracapsular algae. We would not anticipate this consistency in mixed cultures. Therefore we attribute the differences between the intracapsular and cell culture environments to not be attributable to heterogeneous strains, which would likely be far more dramatic (Fang et al. 2012).
4.5 Volunteer vs. cultivated algal associations
The apparent host specificity of tissue invasion by Oophila further suggests that this association is not due to serendipity alone. A recent study found that one algal species, Nanochloris eukaryotum, out of eleven cultures sampled, will voluntarily enter human cells in culture (Black et al. 2014). The cultured alga was extracted from a saltwater fish tank in the former Yugoslavia (Wilhelm and Wild 1982), indicating that its cellular entry was not the product of host-symbiont co-evolution. Similarly, there are sporadic accounts of tissue invasion by otherwise benign Chlorella algae, which can infect sheep or even people (Ramírez-Romero et al. 2015). However, A. maculatum shows no pathological response to its algal symbiont and is apparently uniquely suited for harboring algal cells among the handful of ranid frogs and ambystomatid and hynobiid salamanders that have Oophila algae inside their egg capsules. We recently found that Oophila entry coincides with a stress response by the algae but surprisingly not by the salamander host cells (Burns et al. 2017). The mechanism by which A. maculatum tolerates or even facilitates Oophila entry remains to be determined. There is a community of algae, protists, nematodes, and bacteria within the A. maculatum intracapsular fluid (Gilbert 1942; Hutchison 1971; Kerney et al. 2011). However, to date we have only found tissue entry of a single algal taxon, Oophila amblystomatis, occurring exclusively within a single amphibian host, Ambystoma maculatum.
5 Conclusion
While the data in this paper do show a degree of affinity between A. maculatum and its endogenous algae, the experimental co-culturing of AF6-raised algae with salamander hosts has lower survival and prevalence of tissue entry than those observed in our failed negative control. We are able to conclude from these data that A. gracile is not permissive of tissue entry from either its own symbiotic algae or A. maculatum-derived algae (Kim et al. 2014). Further, the endogenous algae associating with A. maculatum embryos is apparently more beneficial to its hosts and more likely to enter host tissues than cultured algae, suggesting a physiological difference between cultured algae and algal cells found in the wild.
References
Akhtar TA, Orsomando G, Mehrshahi P, Lara-Núñez A, Bennett MJ, Gregory JF, Hanson AD (2010) A central role for gamma-glutamyl hydrolases in plant folate homeostasis. Plant J 64:256–266
Alexa A, Rahnenfuhrer J (2016) topGO: enrichment analysis for gene ontology. R package version 232.0
Bachmann M, Carlton RG, Burkholder J, Wetzel RG (1985) Symbiosis between salamander eggs and green algae: microelectrode measurements inside eggs demonstrate effect of photosynthesis on oxygen concentration. Can J Zool 64:1586–1588
Baldan B, Girard-Bascou J, Wollman FA, Olive J (1991) Evidence for thylakoid membrane fusion during zygote formation in Chlamydomonas reinhardtii. J Cell Biol 114:905–915
Bianchini K, Tattersall GJ, sashaw J, Porteus CS, Wright PA (2012) Acid water interferes with salamander-green algae symbiosis during early embryonic development. Physiol Biochem Zool 85:470–480
Bishop CD, Miller AG (2014) Dynamics of the growth, life history transformation and photosynthetic capacity of Oophila amblystomatis (Chlorophyceae), a green algal symbiont associated with embryos of the northeastern yellow spotted salamander Ambystoma maculatum (Amphibia). Symbiosis 63:47–57
Black CK, Mihai DM, Washington I (2014) The photosynthetic eukaryote Nannochloris eukaryotum as an intracellular machine to control and expand functionality of human cells. Nano Lett 14:2720–2725
Burns J, Zhang H, Hill E, Kim E, Kerney RR (2017) Transcriptome analysis illuminates the nature of the intracellular interaction in a vertebrate-algal symbiosis. eLife 6:e22054
Burr HS (1916) The effects of the removal of the nasal pits in Amblystoma. J Exp Zool 20:27–51
Carrier G, Garnier M, Le Cunff L, Bougaran G, Probert I, De Vargas C, Corre E, Cadoret JP, Saint-Jean B (2014) Comparative transcriptome of wild type and selected strains of the microalgae Tisochrysis lutea provides insights into the genetic basis, lipid metabolism and the life cycle. PLoS One 9:e86889
Cliburn JW, Ward BQ (1963) Occurrence of Oophila amblystomatis (a symbiotic alga) in Ambystoma maculatum of the lower gulf coastal plain. Am Midl Nat 69:508
Dodge JD (1973) The fine structure of algal cells. Academic Press, New York
Fang W, Si Y, Douglass S, Casero D, Merchant SS, Pellegrini M, Ladunga I, Liu P, Spalding MH (2012) Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24:1876–1893
Gibbs M (2003) Axial patterning: using retinoic acid to disrupt homeobox gene expression in axolotls. In: A Practical Guide to Developmental Biology Oxford, England. p. 28–31
Gilbert P (1942) Observations on the eggs of Ambystoma maculatum with especial reference to the green algae found within the egg envelopes. Ecology 23:215–227
Gilbert PW (1944) The alga-egg relationship in Ambystoma maculatum: a case of symbiosis. Ecology 25:366–369
Goff LJ, Stein JR (1976) Preliminary studies on the green alga Oophila in salamander egg masses. J Phycol 12(suppl):23
Guy RD, Vanlerberghe GC, Turpin DH (1989) Significance of phosphoenolpyruvate carboxylase during ammonium assimilation: carbon isotope discrimination in photosynthesis and respiration by the N-limited green alga Selenastrum minutum. Plant Physiol 89:1150–1157
Hale RE, Miller N, Francis RA, Kennedy C (2016) Does breeding ecology alter selection on developmental and life history traits? A case study in two Ambystomatid salamanders. Evol Ecol 30(3):503–517
Hale RE, Kennedy C, Winkelman D, Brown C (2017) An advantage of clear over white egg mass morphs in metabolically demanding microhabitats suggests a role of symbiotic algae in the maintenance of a polymorphism in the spotted salamander (Ambystoma maculatum). Evol Ecol Res 18:637–650
Harrison RG (1969) Harrison stages and description of the normal development of the spotted salamander, Amblystoma punctatum. In: Wilens S (ed) Organization and development of the embryo. Yale University Press, New Haven, p 44–66
Hutchison V (1971) On the Ambystoma egg-alga relationship. Herp Rev 3:82
IUCN, Conservation International, and NatureServe. 2004. Global Amphibian Assessment. IUCN, Conservation International and NatureServe, Washington, DC and Arlington
Jeong SW, Nam SW, HwangBo K, Jeong WJ, Jeong B-R, Chang YK, Park Y-I (2017) Transcriptional regulation of cellulose biosynthesis during the early phase of nitrogen deprivation in Nannochloropsis salina. Sci Rep 7:5264
Kerney R (2011) Symbioses between salamander embryos and green algae. Symbiosis 54:107–119
Kerney R, Kim E, Hangarter RP, Heiss AA, Bishop CD, Hall BK (2011) Intracellular invasion of green algae in a salamander host. Proc Natl Acad Sci U S A 108:6497–6502
Kerney R, Burns JB, Kim E (2017) Chapter 7: investigating mechanisms of algal entry into salamander cells. In: Algal and Cyanobacteria Symbioses. Grube M, Seckbach J, Muggia L (eds) World Scientific Press, London, p 209–239
Kim E, Lin Y, Kerney R, Blumenberg L, Bishop C (2014) Phylogenetic analysis of algal symbionts associated with four North American amphibian egg masses. PLoS One 9:e108915
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lewis LA, Landberg T (2014) Evolutionary diversity of the symbiotic salamander algae, Oophila (Chlorophyta). Unpublished Genbank Submission
Lewis L, Lo C, Urban M, Schwenk K, Xue C, Landberg T (2013) Natural history of the green algae-salamander symbiosis. Phycologia 52(4):62
Lin Y, Bishop CD (2015) Identification of free-living Oophila amblystomatis (Chlorophyceae) from yellow spotted salamander and wood frog breeding habitat. Phycologia 54:183–191
Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG (2009) A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218
Marco A, Blaustein AR (2000) Symbiosis with green algae affects survival and growth of northwestern salamander embryos. J Herp 34:617–621
Mills N, Barnhart M (1999) Effects of hypoxia on embryonic development in two Ambystoma and two Rana species. Physiol Biochem Zool 72:179–188
Moran NA, Yun Y (2015) Experimental replacement of an obligate insect symbiont. Proc Natl Acad Sci U S A 112:2093–2096
Muto K, Nishikawa K, Kamikawa R, Miyashita H (2017) Symbiotic green algae in eggs of Hynobius nigrescens, an amphibian endemic to Japan. Phycol Res 65:171–174
Nema M, Hanson ML, Müller KM (2018) Phylogeny of the egg-loving green alga Oophila amblystomatis (Chlamydomonadales) and its response to the herbicides atrazine and 2,4-D. Symbiosis Online ahead of print. https://doi.org/10.1007/s13199-018-0564-1
Ohkawa H, Hashimoto N, Furukawa S, Kadono T, Kawano T (2011) Forced symbiosis between Synechocystis spp. PCC 6803 and apo-symbiotic Paramecium bursaria as an experimental model for evolutionary emergence of primitive photosynthetic eukaryotes. Plant Signal Behav 6:773–776
Oliver KM, Degnan PH, Burke GR, Moran NA (2010) Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266
Orsomando G, de la Garza RD, Green BJ, Peng M, Rea PA, Ryan TJ, Gregory JF, Hanson AD (2005) Plant gamma-glutamyl hydrolases and folate polyglutamates: characterization, compartmentation, and co-occurrence in vacuoles. J Biol Chem 280:28877–28884
Owen OE, Kalhan SC, Hanson RW (2002) The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277:30409–30412
Pinder A, Friet S (1994) Oxygen transport in egg masses of the amphibians Rana sylvatica and Ambystoma maculatum: convection, diffusion and oxygen production by algae. J Exp Biol 197:1–14
Ramírez-Romero R, Rodríguez-Tovar LE, Nevárez-Garza AM, Lopez A (2015) Chlorella infection in a sheep in Mexico and minireview of published reports from humans and domestic animals. Mycopathologia 169:461–466
Ravanel S, Douce R, Rébeillé F (2011) Chapter 3 - Metabolism of folates in plants, In: Rébeillé F, Douce R (eds) Advances in botanical research. Elsevier, Amsterdam, p 67–106
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Rodriguez-Gil JL, Brain R, Baxter L, Ruffell S, McConkey B, Solomon K, Hanson M (2014) Optimization of culturing conditions for toxicity testing with the alga Oophila sp. (Chlorophyceae), an amphibian endosymbiont. Envir Tox Chem 33:2566–2575
Schultz N (2016) The symbiotic green algae, Oophila (Chlamydomonadales, Chlorophyceae): a heterotrophic growth study and taxonomic history. Master’s Thesis. University of Connecticut. Mansfeld, CT
Sive HL, Grainger RM, Harland RM (2000) Early development of Xenopus laevis. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press
Small DP, Bennett RS, Bishop CD (2014) The roles of oxygen and ammonia in the symbiotic relationship between the spotted salamander Ambystoma maculatum and the green alga Oophila amblystomatis during embryonic development. Symbiosis 64:1–10
Twitty VC (1932) Influence of the eye on the growth of its associated structures, studied by means of heteroplastic transplantation. J Exp Zool 61:333–374
Voolstra CR, Schwarz JA, Schnetzer J, Sunagawa S, Desalvo MK, Szmant AM, Coffroth MA, Medina M (2009) The host transcriptome remains unaltered during the establishment of coral-algal symbioses. Mol Ecol 18:1823–1833
Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, Leong YS, Dong Y, Axford J, Kriesner P, Lloyd AL, Ritchie SA, O'Neill SL, Hoffmann AA (2011) The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476:450–453
Walter W, Sanchez-Cabo F, Ricote M (2015) GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 17:2912–2914
Wantanabe MM, Kawachi M, Hiroki M, Kasai F (2000) NIES collection list of strains. 6 ed. Japan. 1 p
Wilhelm C, Wild A (1982) Growth and photosynthesis of Nanochlorum eucaryotum, a new and extremely small eucaryotic green alga. Z Naturforsch 37:115–119
Acknowledgements
Special thanks to the many Gettysburg College students for their Friday afternoon lab help: Alex Weiss, Mark Manu, Sarah Rivera, Kyle Woodley, and Naufa Armani. This work was supported by an NSF EAGER to R. Kerney, E. Kim and J. Burns, and a Gordon and Betty Moore Foundation grant to R Kerney, J Burns, S Duhamel and D Matus. E Hill and H Zhang were partially supported by an HHMI award to Gettysburg College. Andrew Blaustein, David Craig, Christopher Rombough, Jay Bowerman, Ryan Roysdon, Bridge Joyce, Robin Kodner, Sharyn Marks and especially Kyle Weis helped with the collection of A. gracile embryos. Range map data developed as part of the Global Amphibian Assessment and provided by IUCN-World Conservation Union, Conservation International and NatureServe.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kerney, R., Leavitt, J., Hill, E. et al. Co-cultures of Oophila amblystomatis between Ambystoma maculatum and Ambystoma gracile hosts show host-symbiont fidelity. Symbiosis 78, 73–85 (2019). https://doi.org/10.1007/s13199-018-00591-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-018-00591-2