Abstract
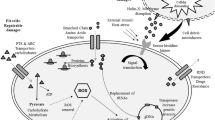
The key functional genes involved in temperature adaption of the Antarctic psychrotrophic bacterium Psychrobacter sp. G. were identified by transcriptomic sequencing. We analyzed the global transcriptional profile of Psychrobacter sp. G under cold stress (0°C) and heat stress (30°C), with the optimal growth temperature 20°C as the control. There were large alterations of the transcriptome profile, including significant upregulation of 11 and 12 transcripts as well as significant downregulation of 47 and 42 transcripts in the cold and heat stress groups, respectively, compared to the control. The expression of various genes encoding enzymes and transcriptional regulators, including PfpI and TetR family transcriptional regulators under heat stress, as well as the expression of DEAD/DEAH box helicase and the IclR family of transcriptional regulators under cold stress, were upregulated significantly. The expression of several genes, most affiliated with TonB-dependent receptor and siderophore receptor, was downregulated significantly under both heat and cold stress. Many of the genes associated with the metabolism of fatty acid and ABC transporters were regulated differentially under different temperature stress. The results of this survey of transcriptome and temperature stress-relevant genes contribute to our understanding of the stress-resistant mechanism in Antarctic bacteria.
Similar content being viewed by others
References
Ashbuner M, Blake J A, Botstein D, et al. 2000. Gene ontology: tool for the unification of biology. Nature Genetics, 25: 25–29
Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics, 29(4): 1165–1188
Bhattacharya C, Wang Xiaolei, Becker D. 2012. The DEAD/DEAH box helicase, DDX11, is essential for the survival of advanced melanomas. Molecular Cancer, 11 (1): 82
Campanaro S, Williams T J, Burg D W, et al. 2011. Temperature-dependent global gene expression in the antarctic archaeon Methanococcoides burtonii. Environmental Microbiology, 13(8): 2018–2038
Che Shuai, Song Weizhi, Lin Xuezheng. 2013a. Response of heatshock protein (HSP) genes to temperature and salinity stress in the Antarctic psychrotrophic bacterium Psychrobacter sp. G. Current Microbiology, 67(5): 601–608
Che Shuai, Song Lai, Song Weizhi, et al. 2013b. Complete genome sequence of Antarctic bacterium Psychrobacter sp. strain G. Genome Announcements, 1(5): e00725–13
Clark M S, Fraser K P P, Burns G, et al. 2008a. The HSP70 heat shock response in the Antarctic fish Harpagifer antarcticus. Polar Biology, 31(2): 171–180
Clark M S, Fraser K P P, Peck L S. 2008b. Antarctic marine molluscs do have an HSP70 heat shock response. Cell Stress and Chaperones, 13(1): 39–49
Clark M S, Fraser K P P, Peck L S. 2008c. Lack of an HSP70 heat shock response in two Antarctic marine invertebrates. Polar Biology, 31(9): 1059–1065
Cornelis P, Bodilis J. 2009. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environmental Microbiology Reports, 1(4): 256–262
Etchegaray J P, Jones P G, Inouye M. 1996. Differential thermoregulation of two highly homologous cold-shock genes, cspA and cspB, of Escherichia coli. Genes to Cells, 1(2): 171–178
Feller G, Gerday C. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nature Reviews Microbiology, 1(3): 200–208
Ferenci T, Spira B. 2007. Variation in stress responses within a bacterial species and the indirect costs of stress resistance. Annals of the New York Academy of Sciences, 1113: 105–113
Filiatrault M J, Stodghill P V, Bronstein P A, et al. 2010. Transcriptome analysis of Pseudomonas syringae identifies new genes, Noncoding RNAs, and antisense activity. Journal of Bacteriology, 192(9): 2359–2372
Folschweiller N, Schalk I J, Celia H, et al. 2000. The pyoverdin receptor FpvA, a TonB-dependent receptor involved in iron uptake by Pseudomonas aeruginosa (Review). Molecular Membrane Biology, 17(3): 123–133
Fonseca P, Moreno R, Rojo F, et al. 2011. Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environmental Microbiology Reports, 3(3): 329–339
Gerday C, Aittaleb M, Bentahir M, et al. 2000. Cold-adapted enzymes: from fundamentals to biotechnology. Trends in Biotechnology, 18(3): 103–107
Hartke A, Frère J, Boutibonnes P, et al. 1997. Differential induction of the chaperonin GroEL and the Co-Chaperonin GroES by heat, acid, and UV-irradiation in Lactococcus lactis subsp. Lactis. Current Microbiology, 34(1): 23–26
Hesami S, Metcalf D S, Lumsden J S, et al. 2011. Identification of coldtemperature-regulated Genes in Flavobacterium psychrophilum. Applied and Environmental Microbiology, 77(5): 1593–1600
Hofmann G E, Buckley B A, Airaksinen S, et al. 2000. Heat-shock protein expression is absent in the Antarctic fish Trematomus bernacchii (family Nototheniidae). Journal of Experimental Biology, 203(15): 2331–2339
Huang Weichun, Marth G. 2008. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Research, 18(9): 1538–1543
Li Jinsong, Bi Yuntian, Dong Cheng, et al. 2011. Transcriptome analysis of adaptive heat shock response of Streptococcus thermophilus. PLoS One, 6(10): e25777
Li Bo, Dewey C N. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12 (1): 323
Lin Xuezheng, Cui Shuoshuo, Xu Guoying, et al. 2010. Cloning and heterologous expression of two cold-active lipases from the Antarctic bacterium Psychrobacter sp. G. Polar Research, 29(3): 421–429
Liu Shenghao, Wang Nengfei, Zhang Pengying, et al. 2013. Next-generation sequencing-based transcriptome profiling analysis of Pohlia nutans reveals insight into the stress-relevant genes in Antarctic moss. Extremophiles, 17(3): 391–403
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–CT method. Methods, 25(4): 402–408
Pearce D A. 2008. Climate change and the microbiology of the Antarctic Peninsula region. Science Progress, 91(2): 203–217
Place S P, Hofmann G. 2005. Constitutive expression of a stress-inducible heat shock protein gene, hsp70, in phylogenetically distant Antarctic fish. Polar Biology, 28(4): 261–267
Place S P, Zippay M L, Hofmann G E. 2004. Constitutive roles for inducible genes: evidence for the alteration in expression of the inducible hsp70 gene in Antarctic notothenioid fishes. American Journal of Physiology, 282(2): R429–R436
Ramos J L, Martínez- Bueno M, Molina-Henares A J, et al. 2005. The TetR family of transcriptional repressors. Microbiology and Molecular Biology Reviews, 69(2): 326–356
Roberta R. 2010. Molecular adaptations in Antarctic fish and bacteria. Polar Science, 4(2): 245–256
Robinson M D, McCarthy D J, Smyth G K. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1): 139–140
Rodríguez-Rojas A, Blázquez J. 2009. The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. Journal of Bacteriology, 191(3): 844–850
Ryabova N, Marchenkov V, Kotova N, et al. 2014. Chaperonin GroEL reassembly: an effect of protein ligands and solvent composition. Biomolecules, 4(2): 458–473
Schuster S C. 2008. Next-generation sequencing transforms today’s biology. Nature Methods, 5(1): 16–18
Sharma C M, Hoffmann S, Darfeuille F, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature, 464(7286): 250–255
Shimizu T, Nakamura A. 2014. Characterization of LgnR an IclR family transcriptional regulator involved in the regulation of Lgluconate catabolic genes in Paracoccus sp. 43P. Microbiology, 160(3): 623–634
Simpson W, Olczak T, Genco C A. 2000. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. Journal of Bacteriology, 182(20): 5737–5748
Song Weizhi, Lin Xuezheng, Huang Xiaohang. 2012. Characterization and expression analysis of three cold shock protein (CSP) genes under different stress conditions in the Antarctic bacterium Psychrobacter sp. G. Polar Biology, 35(10): 1515–1524
Suresh K P, Ghosh M, Pulicherla K K, et al. 2011. Cold active enzymes from the marine psychrophiles: biotechnological perspective. Advanced Biotech, 10(9): 16–20
Thorne M A S, Burns G, Fraser K P P, et al. 2010. Transcription profiling of acute temperature stress in the Antarctic plunderfish Harpagifer antarcticus. Marine Genomics, 3(1): 35–44
Turner J, Colwell S R, Marshall G J, et al. 2005. Antarctic climate change during the last 50 years. International Journal of Climatology, 25(3): 279–294
Wang Jing, Huang Lei, Lu Yimei, et al. 2011. Research on fatty acid compositions of three species of marine bacteria. Periodical of Ocean University of China (in Chinese), 41(S): 252–258
Zeng Yinxin, Chen Bo. 1999. Progress and application prospects in the study on Antarctic cold-adapted microorganisms. Chinese Journal of Polar Research (in Chinese), 11(2): 143–152
Author information
Authors and Affiliations
Corresponding author
Additional information
Foundation item: The National Natural Science Foundation of China under contract No. 41176174; the Chinese Polar Environment Comprehensive Investigation and Assessment Program under contract No. CHINARE 2014-03-05; the Public Science and Technology Funds for Ocean Projects under contract No. 201205020-5.
Rights and permissions
About this article
Cite this article
Wang, Z., Li, Y. & Lin, X. Transcriptome analysis of the Antarctic psychrotrophic bacterium Psychrobacter sp. G in response to temperature stress. Acta Oceanol. Sin. 36, 78–87 (2017). https://doi.org/10.1007/s13131-016-0956-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13131-016-0956-0