Abstract
Feature selection is increasingly important in data analysis and machine learning in big data era. However, how to use the data in feature selection, i.e. using either ALL or PART of a dataset, has become a serious and tricky issue. Whilst the conventional practice of using all the data in feature selection may lead to selection bias, using part of the data may, on the other hand, lead to underestimating the relevant features under some conditions. This paper investigates these two strategies systematically in terms of reliability and effectiveness, and then determines their suitability for datasets with different characteristics. The reliability is measured by the Average Tanimoto Index and the Inter-method Average Tanimoto Index, and the effectiveness is measured by the mean generalisation accuracy of classification. The computational experiments are carried out on ten real-world benchmark datasets and fourteen synthetic datasets. The synthetic datasets are generated with a pre-set number of relevant features and varied numbers of irrelevant features and instances, and added with different levels of noise. The results indicate that the PART approach is more effective in reducing the bias when the size of a dataset is small but starts to lose its advantage as the dataset size increases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
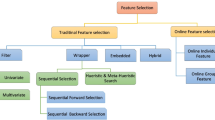
Big data may contain a huge number (from hundreds to millions) of features and often most of the features could be unimportant, irrelevant or redundant, which can cause poor efficiency and/or over-fitting in data analysis and machine learning. Therefore, it is necessary to employ some feature selection methods (FS) to remove irrelevant and redundant features to reduce the complexity of analysis and the generated models and also to improve the efficiency of the whole modelling process [5, 6, 25].
There is, however, a long on-going argument in the field of feature selection about how the data should be used when carrying out feature selection [3, 17, 22, 27]. The central issue is whether all the data, or just some parts of the data should be used in FS before modelling. The ALL approach has become almost a de-facto convention in FS practice primarily because FS is viewed as a mere pre-processing step before analysis, and the ALL approach increases the chance of selecting all the relevant features and then helps to build better models [22, 23]. However, the ALL approach may produce overoptimistic results, as it has used all the data, which means FS has seen the subsets of the data used for later modelling and evaluation. This is called feature subset selection bias. Some studies [3, 17, 22, 27] have discussed this issue and attempted to address it by using the PART approach. Nevertheless, the PART approach may lead to underestimating the relevant features under some conditions [22]. Whilst these studies produced some initial useful insights into the problems, their findings are limited by the facts that these studies were mostly done on rather specific problem domains, such as in genome-wide analysis with wrapper-based feature selection algorithms, on few real-world or artificial datasets with relatively small number of features. Therefore, it is important to evaluate these two approaches systematically and determine their reliability and effectiveness under various circumstances.
The rest of this paper is organized as follows. Section 2 reviews related work. Section 3 describes the methods for the intended research, including the ALL and PART approaches, experiment design and the reliability and effectiveness measures, as well as the selected filters. Section 4 presents the experimental results on real-world benchmark datasets. Section 5 presents the results on synthetic datasets. Summary and Conclusions are presented in Sect. 6.
2 Related work
In recent decades, a few studies [3, 17, 22, 24, 27] have discussed the influence of using FS on the whole dataset and have attempted to solve the selection bias problem by performing FS inside the cross-validation (CV) loop; however, these studies have certain limitations. Ambroise [3] disscused how to correct the selection bias by performing either CV or bootstrap on the selection process. This study used both backward selection with Support Vector Machine (SVM) and forward selection with Linear Discrimination Analysis (LDA) wrapper approaches with only 2 datasets, and did not use filter model. Also, it recommended using tenfolds rather than leaving-one-out for cross-validation. Reunanen [24] studied the FS evaluation method using wrapper models only, but did not address issues specifically relating to the pair-wise comparison of FS algorithms. Lecocke and Hess [17] presented an empirical study in which the PART approach with tenfold CV was applied to filters and wrappers based on genetic algorithms (GA). However, the limitation of their study is that they used just binary classification with microarray data and two FS methods.
Refaeilzadeh et al. [22] attempted to find out which strategy, PART or ALL, is more reliable when conducting pair-wise comparisons of FS algorithms by concentrating on filter models and by using tenfold CV with paired t test. They generated 5 synthetic datasets, but the largest number of features was only 60 and the maximum number of instances was 1000. They explained that there is the potential for bias in both the PART and ALL approaches; with ALL, the FS method has looked at the test set when selecting features, so the accuracy estimate was inflated, whereas with the PART approaches, the FS method uses fewer data than would be available in a real-world experimental setting, which may have led to an underestimated accuracy. The results obtained from their study include: (1) PART and ALL “have different biases, and bias is not a major factor” in determining which one is more truthful in pair-wise comparison; (2) in a greater majority of cases, PART and ALL approaches are not significantly different; (3) the PART approach tends to be more truthful if the two FS methods are performed identically; (4) given two FS methods A1 and A2, for two cases: “(a) A1 is better and (b) A2 is better, if PART is better for case (a), then ALL is better for case (b)” [22]. However, some of their conclusions are not clear, such as they “recommend to run both ALL and PART methods, trust the method indicating that one algorithm is better than the other, and use that the better algorithm to select features using the entire dataset. In the worst case scenario, the selected features will be no worse than the subset selected by the alternative algorithm”. In addition, their study is crucially limited by a fact that they only used synthetic datasets with relatively low dimensions and small number of samples.
3 Methods
3.1 The ALL and PART approaches
The ALL approach uses all the instances in a given dataset in its feature selection step, while the PART approach only uses training instances partitioned from the dataset in feature selection. With the ALL approach, all the data of a dataset are used once in the FS stage, and then the selected features are used as the input variables to generate models, e.g. classifiers, with a common K-fold cross validation procedure as illustrated in Fig. 1a. On the other hand, with the PART approach, as illustrated in Fig. 1b, a dataset is partitioned and some parts are used for FS and also used as the training dataset when inducing classifiers.
This study employs the K-fold cross validation mechanism in the PART approach. It works as follows: K-onefolds are used as the training data for each filter; the selected features are used as the inputs for the classification base learner to build the classifier with the same K-onefolds of the data; then, the remaining fold is used as a validation set to test the classifier. This procedure is repeated round-robin for K times.
3.2 Experiment design
In order to compare the ALL and PART approaches in terms of reliability and effectiveness, several sets of experiments are designed and conducted by using synthetic datasets and real-world benchmark datasets, which extended to our early work [2].
Four common filters (ReliefF, Gain Ratio, CFS and FCBF) are used in parallel as feature selection methods with a hope of avoiding selection bias introduced by individual filters. The detail of these filters and justification of their choice are given in Sect. 3.3
The reliability of these methods is evaluated through measuring the similarity between the selected feature subsets and the desired feature set in synthetic data, or stability when the desired feature set is unknown in real-world benchmark datasets. Average Tanimoto Index (ATI) is used as a stability measure and the Intersystem Average Tanimoto Index (IATI) is modified to measure the similarity. Their details will be described in Sect. 3.4.
The effectiveness of the methods is estimated through measuring the average classification accuracy of the classifiers that are trained with the selected features by the ALL or PART methods. Naïve Bayes classifier (NB) is chosen as the base leaner for the experiments with the synthetic datasets and two more types of classifiers, K-Nearest Neighbours (KNN) and Support Vector Machine (SVM), are used in the experiments on the real-world benchmark datasets in order to evaluate the consistency of classification accuracy, because there is no known answer for the real world datasets.
In general, the main difference between the PART and ALL approaches is in the FS step; The ALL uses all the datasets while the PART uses the training dataset. The experimental process of each approach will be described in detail below.
Firstly, the ALL approach uses the entire dataset in each FS method, and the subsets of the features selected by these FS methods are used as inputs for the classifier. A K-fold (K = 10) cross validation strategy is used when inducting classifiers, and after that the average of the accuracies is calculated as a representation of classification accuracy of the classifiers trained with the features selected by each FS method. Then, each experiment is repeated ten times with different shuffling random seeds in order to assess the consistency of the results. The PART approach uses the training set only (ninefolds) in each FS method, and the subsets produced by these FS methods in each fold are used as the inputs for a base-learner (learning algorithm) to build a classifier, which is then tested on the testing set (remaining fold). This procedure will be repeated 10 times by running filters and the classifier on the training set in each fold, and then testing it on the testing set. After that, we will average the accuracy of ten folds as well as the similarity. Then, each experiment is repeated ten times with different shuffling random seeds in the same way as described above in order to assess the consistency of the results.
The reliability is calculated once for the ALL approach and K times for the PART approach with K-fold cross-validation. The effectiveness of the selected features is measured by the average classification accuracy of the classifier generated with the selected features by the ALL or the PART approaches. The average accuracy as well as the average similarity will be presented in the final results.
In total, 35,200 models were built in our experiments (4 filters × 10 real data sets × 3 classifiers × 2 (PART and ALL) × 10 (runs) × 10 (folds) = 24,000) + (4 filters × 14 synthetic data sets × (PART and ALL) × 10 (runs) × 10 (folds) = 11,200), which is more than any other studies ever did before and therefore our results should be more representative and convincing.
3.3 The filters used for feature selection
This work uses filters as feature selection methods for two reasons: firstly filters are independent of any classifier and generally faster, and secondly there is very limited study found in the literature that has examined the reliability and effectiveness of the PART approach using filter methods. However, filters are designed with different evaluation criteria, which may work well on some datasets but not on others. Therefore, in order to cover a range of type of filters and datasets as wide as possible and to make the investigation more reliable, we follow the categorisation [18] when we select the filters, broadly based on evaluation criteria, e.g., Distance, Information and Correlation. We have therefore chosen ReliefF (from distance measures), Gain Ratio (from information measures), Correlation-based Feature Selection (CFS), and Fast Correlation Based Filter (FCBF). We briefly describe each filter used in this research as follows:
3.3.1 FCBF
Fast correlation based filter [30] starts by sorting features through their correlation with a response using symmetric uncertainty, and optionally removing the bottom of the list according to a pre-specified threshold. Then, the feature that is mostly correlated with the response is selected to add into the minimal subset. After that, all the features that have correlations with the selected feature higher than its correlation with the response are considered redundant and removed. Then the search starts again with the next feature within the remaining feature set.
3.3.2 CFS
Correlation-based feature selection [8] is a simple filtering algorithm that ranks features according to a correlation-based heuristic evaluation function. The key idea of this algorithm is that it employs a heuristic evaluation that assesses the efficacy of individual features in terms of their predicting power for the chosen class. It also assesses how strong the features are inter-correlated. In order to avoid high computational cost, we use liner forward selection (LSF) as a search method together with CSF instead of using Best First Search strategy. LSF is a simple ‘complexity optimization’ of sequential forward selection (SFS). It entails firstly creating a ranking of features and selecting the first K features; then, the SFS algorithm is run over the selected features [7].
3.3.3 ReliefF
This was first proposed by Kira and Rendell [13] and then improved by Kononenko [14] to handle noise and multi-class datasets. The key idea of Relief is that it searches for the nearest neighbours of a sample of each class label, and then weighs the features in terms of how well they differentiate samples for different class labels. This process is repeated for a pre-specified number of instances.
3.3.4 Gain ratio
This is one of the simplest and fastest feature ranking methods. It incorporates ‘split information’ of features into an Information Gain statistic. The split information of a feature is obtained by measuring how broadly and uniformly the data are split. Generally, Gain Ratio evaluates the value of a feature by measuring the gain ratio with respect to the class [21].
Another reason for selecting these four filters is based on the formats of their outputs, which typically fall into two categories: ranking filter (RF) and subset filter (SF). RF evaluates one feature at a time and produces a ranking of all the features in a dataset, whereas SF evaluates subsets of features and outputs the best subset. In order to make our investigation more general, we select two filters from each category: ReliefF and Gain Ratio ∈ RF; FCBF and CFS ∈ SF.
However, when using RF, it is necessary to set a threshold in order to cut off the features that are less relevant from the ranking. Unfortunately, how to set the threshold is a tricky task. Sánchez-Maroño et al. [26] studied the whole ranking process, paying a particular attention to the features that are ranked at top. On the other hand, Belanche and González [4] chose to discard the features that have ranking weights further than two variances from the mean. Others [19] use a threshold defined by the largest gap between two consecutively ranked features. However, in this work we devise a heuristic rule to determine the threshold. After running RFs and SFs, a larger consensus number of features selected by the SFs is taken as a cut-off point for the rankings generated by the RFs. By doing this, we can quickly select the number of features from the rankings.
3.4 Measures of reliability
The reliability of an FS method in this context is measured by computing the degree of similarity between sets of features selected by the PART and ALL approaches, in the case of using a real-world benchmark dataset. In the case of using a synthetic dataset, the degree of similarity is measured between a set of selected features from the ALL or PART approaches and a pre-defined set of desired features. The similarity measure gives us some indication about how far the features selected by the ALL approach are different relatively to the PART approach in each fold and in each run. In addition to the similarity, we measure the stability between the selected features in each fold with the PART approach in order to quantify that how different training sets may affect the feature preference.
The stability of FS was defined by Han and Yu [9] as the robustness of the result of an FS algorithm to variations in the training set. The stability measure can be used in different situations; it is necessary for evaluating different FS methods in performance comparison. Also, it can be used for the internal validation of FS algorithms that take into account stability [10]. Measuring reliability requires a similarity measure for FS results. There are three types of FS representation methods: subset of features, ranking vector and weighting score vector. In this work, we focus on subset of features because some of the filters used in this study produce subsets of features.
There are quite a few similarity measures available for comparison of sets, as reviewed by He and Yu [10]. The measures presented in Křížek et al. [15] and Kuncheva [16] are both subset-based, but they can only be used on subsets of equal cardinality. However, in our research, the subset cardinality is not equal, so we use the Average Tanimoto Index (ATI), which allows us to use subsets of unequal size. It is defined below:
Average Tanimoto Index (ATI) [28] is computed over all subset pairs, and then averaged. It is a continuous value from [0, 1], with 0 representing an empty intersection between subsets \(X_{i}\), \(X_{j}\) and 1 representing that all subsets obtained from n runs are identical:
where X is a set of all features, \(X_{s}\) is a set of k features selected from k-fold runs. \(X_{i}\) and \(X_{j} \in X_{s}\), and k is the number of folds. Similarity measures \(Sim\) between two sets X i , X j [12] is defined as:
All the measures discussed above consider intra-measures, which are used for evaluating the internal stability of one FS process, as in the PART approach. We cannot use it for the ALL approach because the entire dataset is used and there is no change in the dataset during each run, so the same feature subset is produced in each run when the set-ups for each filter is fixed. Also, with intra-measures we cannot compare the subsets produced by each filter with the optimal features because there is no optimal answer (relevant features) in real-world dataset, which motivated us for generating the synthetic dataset. Therefore, we include a second measure in our investigation, called inter-measures, in order to compare the result of each approach (ALL, PART) with the relevant features on synthetic data, and also compare the results of the ALL and PART approaches on a real-world benchmark dataset. The inter-measures should provide information that is complementary to the intra-measures. Therefore, the following inter-measure is defined as an equivalence to intra-measure, based on the same principle [28].
The original Inter-method Average Tanimoto Index \(({\text{IATI}}_{R} )\) between two methods \(X_{s}^{1}\) and \(X_{s}^{2}\) is defined as:
where \(X_{s}^{1}\), \(X_{s}^{2}\) are the two sets of the feature subset selected by the two methods respectively; which takes values from [0, 1] with 0 indicating an empty intersection between any pair of subsets, and 1 indicating that all subsets in both methods \(X_{s}^{1}\) and \(X_{s}^{2}\) are identical. Also, \(k_{1 } \;and\;k_{2 }\) are the number of folds used to generate \(X_{s}^{1}\), \(X_{s}^{2}\) [28].
However, we found that this definition is highly affected by the size of X, which leads to decreasing the similarity when the number of features increases. Therefore, we modify it by removing |X| to avoid this drawback. It is now defined as follows:
In fact, by using a real-world dataset, any similarity measure including the ones described above can only indicate the similarity degree between the ALL and PART approaches, but cannot tell which one is better when they are dissimilar. So, we need to evaluate how effective they are by measuring the average classification accuracy as described below.
3.5 Effectiveness measures of feature selection
The effectiveness of the selected features is measured by the average classification accuracy of the classifiers that are generated with the features selected by the ALL or the PART approaches. Generally, classification performance is dependent of the types of classifiers used, even under the same conditions, with the same subset of features and samples, and same training procedure. To avoid any bias produced by classifiers of the same type when comparing the effectiveness of both the ALL and PART approaches, in our experiments, we use three types of classifier: NB (Naïve Bayesian) [11], KNN (k Nearest Neighbours) [1] and SVM (Support Vector Machine) [20]. These three algorithms have been chosen because they represent three quite different approaches in machine learning, and they do not contain any embedded feature selection mechanism; also, they are commonly used in data mining practice.
The statistical significance of the results of multiple runs for each experiment is calculated and compared between accuracies with Student’s t-test and Wilcoxon signed rank test [29] at a significance level of 0.05.
4 Experiments with real-world benchmark data
4.1 The real-world benchmark datasets
Ten real-world benchmark datasets from different domains are used in our experiments. Six of them (Zoo, Dermatology, Promoters, Splice, Multi-feature-factors and Arrhythmia) are from the UCI Machine Learning Repository,Footnote 1 two others (Colon and Leukaemia) are from the Bioinformatics Research Group,Footnote 2 and the final two (SRBCT and Ovarian) are from the Microarray Datasets website.Footnote 3
Table 1 summarizes general information on these datasets. Note that these datasets differ greatly in sample size, S, ranging from 62 to 3191 and number of features, NT, ranging from 17 to 15,154. Also, they include binary-class and multi-class classification problems; this should provide a wider basis for testing and should be well suited to the FS methods under differing conditions.
4.2 The results
This section presents the summarised results from the four filters over ten real-world benchmark datasets. The behaviour of the FS method will be evaluated according to similarity between the PART and ALL approaches, stability with the PART approach, and the classification accuracy obtained by the NBC, KNN and SVM models.
4.2.1 Results of comparing reliability
The similarity measure will give us some indication about how far the features selected by the ALL approach are different from those by the PART approach in each fold and in each run.
Figure 2 shows the similarity measures of IATI with the features selected by the 4 filters, comparing the PART and ALL approaches, which on average scored 0.60, 0.66, 0.58 and 0.76, respectively. In light of the results shown in Fig. 2, the similarity between the PART and ALL approaches is indeed affected by the type of filter. As we can see, the Gain Ratio filter delivered higher similarities between these two approaches, when compared with the other filters, especially ReliefF.
Additionally, the similarity between the PART and ALL approaches is affected by the type of dataset. As can be seen, the last 6 datasets have less similarity between the PART and ALL approaches than the first four datasets. This is because they are microarray datasets with quite large numbers of features and very small number of samples. Also, the M-feat-factor and Arrhythmia datasets have less similarity than the first four datasets and this may be because their numbers of class labels are large (10 with the M-feat-factor and 13 with Arrhythmia).
However, the similarity measure with these real-world benchmark datasets can only indicate the extent of similarity between the ALL and PART approaches, it cannot tell which one is better when they are dissimilar. Thus, it is necessary to evaluate how effective they are by measuring their average classification accuracy.
Figure 3 shows different stability values of each filter on the same dataset when the PART approach is used in FS. It is apparent that some filters are more stable than others as we can see, ReliefF has a higher average stability for all the datasets, scoring an average 0.78, and after that, Gain Ratio scored 0.73, which indicates that ranking filters are more stable when the PART approach is used in FS than the SF. In contrast, SF (FCBF and CFS) is unstable as we can see the scores on average of 0.55 and 0.60, respectively. Also, the stability is affected by the different datasets; as we can see, the first five datasets are more stable than the last five.
Similar to the similarity measures (IATI), stability measures (ATI) can only indicate which filters are more stable than others but they cannot tell which one is more accurate in selecting the relevant features, until we evaluate how effective they are by measuring the average classification accuracy.
4.2.2 Results of comparing effectiveness
The figures given in this section show the average accuracy of the NB, KNN and SVM models on the ten real-world benchmark datasets; each value presented in the figures is the average over ten runs of ten-fold cross-validation outcomes using the ALL and PART approaches.
Figure 4 shows the results on the ten datasets with the NB classifiers and the accuracy comparison between the PART and ALL approaches trained with the features selected by the four filters. The PART and ALL approaches produce different results to some extent, affected by the type of filter and the type of data. The accuracy by using the PART approach decreases on average by −1.292, −1.219, −0.689 and −0.731, respectively, relative to the ALL approach. Figure 5 shows the difference between the average accuracies of the NB classifiers trained by the ALL and PART approaches; if the difference is positive, it means that ALL has a higher accuracy than PART, hence is better; while if it is negative, it means that the PART approach has a higher accuracy. As we can see, the ALL approach has the higher accuracy in the majority, relative to the PART approach. Furthermore, microarray datasets (Colon to Leukaemia) in particular, exhibit a significant decline with the PART approach in most of the filter methods.
The results in Fig. 6 show the performance of the KNN (k = 1) classifiers; the accuracy by using the PART approach decreases on average by −1.007, −1.064, −1.233 and +0.196 respectively, relative to the ALL method. Figure 7 shows the difference between the average accuracies of the KNN classifiers trained by the ALL and PART approaches. ReliefF has the highest decline with the PART approach, followed by the other FS models, while Gain Ratio increases the accuracy by using the PART approach. Moreover, the degree of significant change in the accuracy between the PART and ALL approaches differs from one classifier to another as well as from one filter to another.
Figure 8 shows the accuracies of the SVM models and the comparisons between the filers. It can be observed that the accuracy by using the PART approach decreases on average relative to the ALL approach by −0.94, −0.828, −0.821 and −0.396, respectively. Figure 9 shows the difference between the average accuracies of the SVM classifiers trained by the ALL and PART approaches. As we can see, the ALL approach has the highest accuracy in the majority of situations. Furthermore, the classification accuracy of SVM models on the microarray datasets (Colon to Leukaemia) in particular exhibits significant declines with the PART approach in most of the filter methods.
Table 2 summarizes the wins/ties/losses in accuracy comparing the PART approach with the ALL approach over all the datasets on three classifiers by using the Student’s paired two tailed t test (with a significance level of 0.05). The results shown in the table clearly reveal that the PART approach has significantly more losses in the greater majority of cases with FCBF, CFS and ReliefF, which could be an indication that the ALL approach indeed produced some degree of the so-called selection bias. On the other hand, with Gain Ratio, the PART and ALL approaches are not significantly different in most cases.
The results in Table 3 summarize the wins/ties/losses in accuracy comparing the PART approach with the ALL approach over all the datasets on three classifiers. The Wilcoxon signed ranks test (with a significance level of 0.05) is used to test the significance of the ranking differences among them. The similar patterns to the ones appeared in Table 2 can also be observed, which clearly reveal that the PART approach has significantly more losses in the greater majority of cases with FCBF, CFS and ReliefF. This may imply that the ALL approach indeed produced some degree of the so-called selection bias. On the other hand, with Gain Ratio, the PART and ALL approaches are not significantly different in most cases.
Also, we are interested in this section in understanding the relationship between the level of similarity vis-à-vis PART and ALL and the level of change in classification accuracy between them.
In light of the results shown in Fig. 10, the differences between the ALL and PART approaches are affected by 3 factors: dataset, filter type and classifier types. Thus, by changing one of these factors, the result between ALL and PART will also change.
The difference (∆acc) between the average accuracies of the three classifiers trained by the ALL and PART approaches as well as the averages for IATI and ATI. ∆acc = acc (ALL) − acc (PART), represents the difference between the average accuracies of the three classifiers trained by the ALL and PART methods
The similarity measure, IATI, can only indicate the degree to which the ALL and PART approaches are similar, but cannot tell which one is better when they are dissimilar. The stability measure ATI can only indicate with the PART approach how far each filter is stable by using different datasets. Moreover, the accuracy of the three classifiers provides different patterns when using the ALL and PART approaches. Although the above results demonstrate that the accuracy through using the PART approach is lower than through using the ALL approach in most cases, and that the level of similarity between the PART and ALL approaches differs from one FS method to another, these results do not give us a clear picture to determine which approach provides less bias and is more reliable to use. Also, we do not know which approach helps us in selecting the more relevant features, as we applied the experiment on real-world benchmark data without knowing the most relevant features. Therefore, in the next section, we will use the generated synthetic data in order to apply the experiment on a dataset in which we know the relevant features in advance; this will help us to answer the above questions.
5 Experiment with synthetic data
5.1 Synthetic datasets
In principle, generating and using synthetic data is considered to be a useful strategy for testing the effectiveness of FS methods for the following reasons [4]:
-
1.
Knowing the optimal features in synthetic data in advance is the most important advantage. Then, the performance of a FS algorithm can be easily evaluated by computing the degree of matching between the features selected by that algorithm and the known optimal solution.
-
2.
Being able to conduct the investigations in a systematic way, by systematically varying the experimental conditions, such as changing the ratio between the number of samples and number of features, or adding more irrelevant features or noise to the data.
In practice, this strategy facilitates studying key underlying issues and quantitatively assessing the performance of the existing algorithms.
The datasets generated for this study are intended to represent different aspects, such as varying (a) the number of irrelevant features, (b) the number of instances, and (c) levels of noise in the data. These factors can make the FS task very difficult.
The synthetic datasets are generated based on a linear function defined by Eq. (5) and all features have continuous values (even the response variable). However, in order to use these datasets in the classification problem, the response variable is converted to binary. The reason for using liner synthetic datasets is just to simplify the problem and to focus more precisely on our investigation.
The following steps were taken to generate these datasets, where \(N_{R}\) represents the number of relevant features, \(N_{I}\) the number of irrelevant features, \(N_{T}\) the number of total features, S the number of instances, and y c the response variable.
-
Step 1 Generate random matrix (N T , S) of S samples with \(N_{T}\) features, with a given mean μ and a standard deviation σ. Then we expand this matrix by increasing \(N_{T}\) and S.
-
Step 2 Select N R as relevant features, and generate their coefficient \(\upbeta_{i}\) and multiply \(N_{R} \left( {x_{1} , \ldots ,x_{{N_{R} }} } \right)\) with the β value.
$${\beta = }\{\upbeta_{1 } ,\upbeta_{2 } , \ldots ,\upbeta_{{N_{R} }} \} ,\quad {\text{S}}.{\text{T}}:\sum\limits_{i = 1}^{{N_{R} }} {\upbeta_{i} } = 1$$ -
Step 3 Compute the response variable y c by the following equation:
$$y_{c} = \mathop \sum \limits_{i = 1}^{{N_{R} }}\upbeta_{i} x_{i} + \mathop \sum \limits_{j = 1}^{{N_{I} }} \gamma_{j} x_{j}$$(5)where, \(\upgamma_{j} (j = 1, \ldots ,N_{I} )\), are the coefficients of irrelevant features and can be set to zero, so that \(N_{I}\) features make no contribution the response and thus become irrelevant.
-
Step 4 Convert the response variable y c from continuous to binary by:
$$y = \left\{ {\begin{array}{*{20}c} {0,} & {y_{c} < \bar{y}} \\ {1,} & {y_{c} \ge \bar{y}} \\ \end{array} } \right.$$(6)where
$$\bar{y} = \frac{{\sum {y_{{c{\text{i}}}} } }}{S}$$(7)In this study, N R is set to be 10 for all the synthetic datasets, then the response values are computed by the equations and conditions as follows:
$$y_{c} =\upbeta_{1 } x_{1} +\upbeta_{2 } x_{2} + \cdots +\upbeta_{10} x_{10}$$(8)\(\upbeta_{i + 1} =\upbeta_{i} + {\Delta \beta ,}\quad {\text{i = 1 to 10}} .\) where \(\upbeta_{1} = 0.01 \;and \;{\Delta \beta } = 0.02\)
In order to simulate a characteristic of real datasets, which usually have different degrees of noise, we inject class noise into 3 datasets (S2, S5 and S8) with differing rates. The first parameter, denoted p (p = 5, 10 %), is used to determine the number of samples injected by noise. The second parameter, denoted ε, which is a random number varying between \(\varepsilon = - 0.1 \to 0.1\), represents the magnitude of noise level injected to response variable.
Table 4 lists the synthetic datasets generated for a classification problem with a fixed number (10) of relevant features, varied numbers of irrelevant features, the number of instances, and levels (5, 10 %) of noise.
5.2 The results
This section presents the results generated after applying four filters over fourteen synthetic datasets. The behaviour of the FS methods will be evaluated in terms of the similarity computed between the features selected with the PART and ALL approaches and the optimal set of the features, the stability with the PART method, and the classification accuracy obtained by the NB classifier.
5.2.1 Results of comparing reliability
Figure 11 gives the summary of the results for reliability measured by similarity measure IATI for all the synthetic datasets. It shows that the PART and ALL approaches are mostly similar with all the filters on almost all of the datasets, except for a few cases, as shown in Fig. 12. One case is S4-PART versus S4-ALL, where the number of instances is small (100) and the number of the irrelevant features is relatively very large (990), 99 times larger than the number of the relevant features. Another case is when the noise level is increased to 10 %; the PART approach appeared to be slightly worse than the ALL approach.
In addition, in Fig. 12, it is worth noting how the similarity in S2Noise5 and S2Noise10 datasets (with both the PART and ALL approaches) decreases quite significantly from 0.81 to 0.64, when the noise level increases from 5 to 10 %, while there is almost no difference between S5 and S8 with 10 % noise relative to 5 % noise. Therefore, we can say that datasets with small numbers of samples (as S2) can be easily affected by noise, more so than data with large numbers of samples. Accordingly, we can note that the number of samples plays the most important role. As we can observe, if the number of samples is small, it will be hard for any of the filters to select a large number of relevant features; moreover, we notice an increasing tendency to select more irrelevant features. Additionally, the results indicate that increasing the number of irrelevant features in the dataset can have a quite strong adverse effect on the performance of filters, as it also increases the chance of choosing irrelevant features.
Figure 13 shows the results of ATI measures in the PART approach. We clearly notice the big differences in stability between the datasets, primarily due to the increases in the number of samples and irrelevant features. We observe a higher stability in S3 and S6 because of the large number of samples (10,000). Besides, we can clearly see (1) that increasing irrelevant features causes a decrease in stability, as in datasets S4, S7 and S8; and (2) a greater decrease in stability because of the increase in noise from 5 to 10 %.
5.2.2 Results of comparing effectiveness
Figure 14 shows the average test accuracy of the NB classifiers trained with the features selected by both approaches. The best classification accuracy was obtained by S3-PART as well as S3-ALL (S = 10,000 and N I = 90). The worst classification accuracy as well as the lowest similarity were obtained by S7-PART (S = 100 and N I = 9990). Within these two datasets, we can see various classification accuracy results, varying based on two factors in general: the number of samples and the number of irrelevant features. From Fig. 15 we can clearly observe that the ALL approach has a higher accuracy than the PART approach on the datasets with small numbers of samples (as in S1, S4 and S7) and the difference between the ALL and PART approaches increases as N I increases. The NB classifiers trained with S7-ALL in particular greatly outperformed those trained with S7-PART, by about 47.2 % on average in terms of accuracy, while both approaches give very low similarity, as seen in Fig. 11; this case simulates the special characteristics of microarray datasets, i.e. having a very large feature dimensionality and a very small number of samples. On the other hand, the PART and ALL approaches obtained similar accuracy on the remaining datasets, which have medium or large numbers of samples.
Further observations reveal that NB classifiers trained with datasets S2, S5 and S8 (without any noise) achieved higher accuracies than those of the classifiers for the datasets with 5 and 10 % added noise. Moreover, Fig. 15 shows a little decrease in the accuracy by increasing the noise rate. For example, S2Noise5-ALL scored 93.04 % with most of the filters, while S2Noise10-ALL scored 91.19 % with all the filters.
6 Summary and conclusions
In this paper, the differences between the PART and ALL approaches have been investigated in terms of similarity, stability and classification accuracy on 10 real-world benchmark datasets and 14 synthetic datasets generated.
The findings can be summarised as follows. Firstly, the PART and ALL approaches produce no obvious difference in terms of accuracy and similarity on the real-world benchmark and synthetic datasets with large numbers of samples, such as S3, S6 and Splice, and also have high stability. Secondly, they also demonstrate no obvious differences in terms of accuracy and similarity with the IATI measure on those datasets with medium numbers of samples, such as S2, S5 (S = 1000) and Dermatology, unless the datasets with a large number of irrelevant features, such as S8 and M-feat-factors. Thirdly, these two approaches are demonstrated to have only small differences in accuracy and similarity, and also have high stability on those datasets with small numbers of samples and very small numbers of features, such as Zoo (N T = 17) and Promoters (N T = 57). Finally, they show clear differences in accuracy on the datasets with small numbers of samples, such as S1, S4, S7 (S = 100), Colon and Leukaemia, which indicates that the ALL approach achieves higher accuracy than the PART approach, although the similarity and stability results are still low in both the methods.
In addition, the experiment results lead to some more general conclusions as follow:
-
1.
The number of samples plays a major role in the performance of FS. Whenever the number of samples increases, this leads to the FS method selecting more relevant features and discarding irrelevant ones. Also, it leads to increasing the similarity and stability in addition to the classification accuracy.
-
2.
The number of irrelevant features is an important factor in the performance of FS, as increasing the number of irrelevant features in the dataset disrupts the FS process and increases the possibility of choosing irrelevant features; in addition, it reduces the similarity, stability and classification accuracy.
-
3.
Finally, the level of noise is another important factor influencing the FS process in which increases the chances of choosing irrelevant features as well as decreasing the similarity, stability and classification accuracy.
In conclusion, when the dataset contains a large number of samples there is no noticeable difference between these two approaches in terms of reliability and effectiveness. When the dataset is small, the ALL and PART approaches have almost similar reliability. However, there is a clear difference in terms of their effectiveness, that is, the ALL approach achieves a higher accuracy than the PART approach, which indicates that the accuracy estimate is possibly overstated and that bias has occurred. Therefore, the PART approach can prevent bias to some extent, although its superiority decreases with increasing sample sizes.
References
Aha DW, Kibler D, Albert MK (1991) Instance-based learning algorithms. Mach Learn 6:37–66
Aldehim G, Wang W (2014) Reliability and effectiveness of cross-validation in feature selection. In: Bramer M, Petridis M (eds) Research and development in intelligent systems XXXI. Springer, pp 179–184
Ambroise C (2002) Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci 99:6562–6566. doi:10.1073/pnas.102102699
Belanche L, González F (2011) Review and evaluation of feature selection algorithms in synthetic problems. arXiv:11012320
Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A (2013) A review of feature selection methods on synthetic data. Knowl Inf Syst 34:483–519
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40:16–28
Gutlein M, Frank E, Hall M, Karwath A (2009) Large-scale attribute selection using wrappers. Paper presented at the computational intelligence and data mining
Hall MA (1999) Correlation-based feature selection for machine learning. The University of Waikato, Hamilton
Han Y, Yu L (2012) A variance reduction framework for stable feature selection. Stat Anal Data Min 5:428–445
He Z, Yu W (2010) Stable feature selection for biomarker discovery. Comput Biol Chem 34:215–225. doi:10.1016/j.compbiolchem.2010.07.002
John GH, Langley P (1995) Estimating continuous distributions in Bayesian classifiers. In: Proceedings of the eleventh conference on uncertainty in artificial intelligence, San Francisco, CA, USA. Morgan Kaufmann, pp 338–345
Kalousis A, Prados J, Hilario M (2007) Stability of feature selection algorithms: a study on high-dimensional spaces. Knowl Inf Syst 12:95–116
Kira K, Rendell LA (1992) The feature selection problem: traditional methods and a new algorithm. In: Proceedings of the tenth national conference on artificial intelligence, San Jose, California. AAAI Press, pp 129–129
Kononenko I (1994) Estimating attributes: analysis and extensions of RELIEF. In: Proceedings of European conference on machine learning Catania, Italy. Springer, pp 171–182. doi:10.1007/3-540-57868-4_57
Křížek P, Kittler J, Hlaváč V (2007) Improving stability of feature selection methods. In: Computer analysis of images and patterns. Springer, pp 929–936
Kuncheva LI (2007) A stability index for feature selection. In: Proceedings of the 25th IASTED international multi-conference: artificial intelligence and applications, ACTA Press, pp 390–395
Lecocke M, Hess K (2006) An empirical study of univariate and genetic algorithm-based feature selection in binary classification with microarray data. Cancer Inform 2:313–327
Liu H, Yu L (2005) Toward integrating feature selection algorithms for classification and clustering. Knowl Data Eng 17:491–502
Mejía-Lavalle M, Sucar E, Arroyo G (2006) Feature selection with a perceptron neural net. In: Proceedings of the international workshop on feature selection for data mining, pp 131–135
Platt JC (1999) Fast training of support vector machines using sequential minimal optimization. In: Smola AJ (ed) Advances in Kernel methods. MIT Press, Cambridge, pp 185–208
Quinlan JR (1993) C4 5: programs for machine learning, vol 1. Massachusetts, Morgan kaufmann
Refaeilzadeh P, Tang L, Liu H (2007) On comparison of feature selection algorithms. In: Proceedings of AAAI workshop on evaluation methods for machine learning II, pp 34–39
Refaeilzadeh P, Tang L, Liu H (2009) Cross-validation. In: Encyclopedia of database systems, Springer, pp 532–538
Reunanen J (2003) Overfitting in making comparisons between variable selection methods. J Mach Learn Res 3:1371–1382
Saeys Y, Inza I, Larranaga P (2007) A review of feature selection techniques in bioinformatics. Bioinformatics 23:2507–2517. doi:10.1093/bioinformatics/btm344
Sánchez-Maroño N, Alonso-Betanzos A, Tombilla-Sanromán M (2007) Filter methods for feature selection–a comparative study. In: Intelligent data engineering and automated learning-IDEAL 2007, Springer, pp 178–187
Singhi SK, Liu H (2006) Feature subset selection bias for classification learning. In: Proceedings of the 23rd international conference on machine learning ACM, pp 849–856
Somol P, Novovicova J (2010) Evaluating stability and comparing output of feature selectors that optimize feature subset cardinality. Pattern Anal Mach Intell 32:1921–1939. doi:10.1109/TPAMI.2010.34
Wilcoxon F (1945) Individual comparisons by ranking methods. Biom Bull 1:80–83. doi:10.2307/3001968
Yu L, Liu H (2004) Efficient feature selection via analysis of relevance and redundancy. J Mach Learn Res 5:1205–1224
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Aldehim, G., Wang, W. Determining appropriate approaches for using data in feature selection. Int. J. Mach. Learn. & Cyber. 8, 915–928 (2017). https://doi.org/10.1007/s13042-015-0469-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-015-0469-8