Abstract
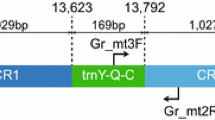
Hypancistrus zebra is an ornamental fish endemic to the Xingu river, Amazon basin, which is under the impact zone of the world’s fourth-largest hydroelectric dam. Illegal capture is another threat to this species. Despite its critical conservation status only two nucleotide sequences from this fish are publicly available on GenBank and on the BOLD System. Here, the nearly complete mitochondrial genome of H. zebra is described; totalizing 16,330 nucleotides, including the complete sequences of the two ribosomal RNA subunits, the 22 transfer RNAs and the 13 protein coding genes. The mitochondrial genome was assembled from transcriptome data, from seven organs, two individual fish, and to an average sequencing depth of 7245x. Seven nucleotide differences were found between the individual fish sequenced here-in, while four were detected among these individuals and the unique mitochondrial sequence publicly available. Additionally, 21 heteroplasmic sites were found among seven organs. This is the first quasi complete mitochondrial genome of a species belonging to the Peckoltia Clade, a Loricariidae phylogenetically problematic tribe. This genetic resource will be valuable in the efforts to elucidate the phylogenetic relationships among the Peckoltia Clade species, as well as it shall subsidize future practices aiming the conservation of this endangered species.

Similar content being viewed by others
References
Armbruster JW (2004) Phylogenetic relationships of the suckermouth armoured catfishes (Loricariidae) with emphasis on the Hypostominae and the Ancistrinae. Zool J Linn Soc 141:1–80. doi:10.1111/j.1096-3642.2004.00109.x
Armbruster JW (2008) The genus Peckoltia with the description of two new species and a reanalysis of the phylogeny of the genera of the Hypostominae (Siluriformes: Loricariidae). Zootaxa 1822:1–76
Armbruster JW, Werneke DC, Tan M (2015) Three new species of saddled loricariid catfishes, and a review of Hemiancistrus, Peckoltia, and allied genera (Siluriformes). Zookeys 123:97–123. doi:10.3897/zookeys.480.6540
Bernt M, Donath A, Jühling F et al (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69:313–319. doi:10.1016/j.ympev.2012.08.023
Buckup PA, Menezes NA (2007) Catálogo das Espécies de Peixes de Água Doce do Brasil.
Covain R, Fisch-Muller S, Oliveira C et al (2016) Molecular phylogeny of the highly diversified catfish subfamily Loricariinae (Siluriformes, Loricariidae) reveals incongruences with morphological classification. Mol Phylogenet Evol 94:492–517. doi:10.1016/j.ympev.2015.10.018
Cramer CA, Bonatto SL, Reis RE (2011) Molecular phylogeny of the Neoplecostominae and Hypoptopomatinae (Siluriformes: Loricariidae) using multiple genes. Mol Phylogenet Evol 59:43–52. doi:10.1016/j.ympev.2011.01.002
Iwasaki W, Fukunaga T, Isagozawa R et al (2013) Mitofish and mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol 30:2531–2540. doi:10.1093/molbev/mst141
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323. doi:10.1186/1471-2105-12-323
Lujan NK, Armbruster JW, Lovejoy N, López-fernández H (2015) Multilocus molecular phylogeny of the suckermouth armored catfishes (Siluriformes: Loricariidae) with a focus on subfamily Hypostominae. Mol Phylogenet Evol 82:269–288. doi:10.1016/j.ympev.2014.08.020
Moreira DA, Furtado C, Parente TE (2015) The use of transcriptomic next-generation sequencing data to assemble mitochondrial genomes of Ancistrus spp. (Loricariidae). Gene 573:171–175. doi:10.1016/j.gene.2015.08.059
Moreira DA, Buckup PA, Andrade PCC et al (2016a) The complete mitochondrial genome of Corydoras nattereri (Callichthyidae: Corydoradinae). Neotrop Ichthyol 14:e150167. doi:10.1590/1982-0224-20150167
Moreira DA, Magalhaes MGP, de Andrade PCC et al (2016b) An RNA-based approach to sequence the mitogenome of Hypoptopoma incognitum (Siluriformes: Loricariidae). Mitochondrial DNA Part A, DNA mapping, Seq Anal 27:3784–3786. doi:10.3109/19401736.2015.1079903
Ray CK, Armbruster JW (2016) The genera Isorineloricaria and Aphanotorulus (Siluriformes: Loricariidae) with description of a new species. Zootaxa 4072:501–539. doi:10.11646/zootaxa.4072.5.1
Rosa RS, Lima FCT (2008) Peixes. In: Machado ABM, Drummond GM, Paglia AP (eds) Livro Vermelho da Fauna Brasileira Ameaçada de Extinção, 2nd edn. Fundação Biodiversitas, BRASÍLIA, pp 9–278
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi:10.1093/bib/bbs017
Acknowledgements
Authors are grateful to Dr. Jansen Zuanon from the Instituto Nacional de Pesquisas da Amazônia (INPA) for the donation of Hypancistrus zebra specimens (Voucher INPA 46655) used in this study. T.E.P. thanks Dr. Adalberto L. Val, Nazaré Paula and the entire ADAPTA and LEEM staffs for their hospitality and assistance during his visit to INPA. This work was supported by the U.S. Agency for International Development under Grants PGA-2000003446 and PGA-2000004790; and by the Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior (CAPES) from the Brazilian Government for a master fellowship for M. G. P. M., a doctoral fellowship for D. A. M., and a postdoctoral fellowship to T. E. P.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Magalhães, M.G.P., Moreira, D.A., Furtado, C. et al. The mitochondrial genome of Hypancistrus zebra (Isbrücker & Nijssen, 1991) (Siluriformes: Loricariidae), an endangered ornamental fish from the Brazilian Amazon. Conservation Genet Resour 9, 319–324 (2017). https://doi.org/10.1007/s12686-016-0645-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-016-0645-5