Abstract
Tuberculosis (TB) is a chronic infectious lung disease, which caused the death of about 1.5 million people in 2020 alone. Therefore, it is important to detect TB accurately at an early stage to prevent the infection and associated deaths. Chest X-ray (CXR) is the most popularly used method for TB diagnosis. However, it is difficult to identify TB from CXR images in the early stage, which leads to time-consuming and expensive treatments. Moreover, due to the increase of drug-resistant tuberculosis, the disease becomes more challenging in recent years. In this work, a novel deep learning-based framework is proposed to reliably and automatically distinguish TB, non-TB (other lung infections), and healthy patients using a dataset of 40,000 CXR images. Moreover, a stacking machine learning-based diagnosis of drug-resistant TB using 3037 CXR images of TB patients is implemented. The largest drug-resistant TB dataset will be released to develop a machine learning model for drug-resistant TB detection and stratification. Besides, Score-CAM-based visualization technique was used to make the model interpretable to see where the best performing model learns from in classifying the image. The proposed approach shows an accuracy of 93.32% for the classification of TB, non-TB, and healthy patients on the largest dataset while around 87.48% and 79.59% accuracy for binary classification (drug-resistant vs drug-sensitive TB), and three-class classification (multi-drug resistant (MDR), extreme drug-resistant (XDR), and sensitive TB), respectively, which is the best reported result compared to the literature. The proposed solution can make fast and reliable detection of TB and drug-resistant TB from chest X-rays, which can help in reducing disease complications and spread.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tuberculosis (TB) is a contagious disease and the leading infectious disease-related cause of death [1]. TB can be cured if diagnosed early and treated properly [2]. Chest X-rays (CXRs) are routinely utilized for pulmonary tuberculosis detection and screening [3, 4]. Chest radiographs are analyzed in clinical practice by trained medical doctors in TB diagnosis. However, this is prone to subjective evaluation, expert-dependent, and sometimes inefficient process. Subjective discrepancies in radiograph-based illness diagnosis are unavoidable [5, 6]. CXR images of TB patients are sometimes confused with other lung abnormalities of similar patterns [7, 8]. This leads to incorrect diagnosis and therapeutic treatment which worsen the disease of the patients. Moreover, radiologists are in short supply in low-income countries, particularly in countryside areas. Computer-assisted diagnostic (CAD) systems that analyze chest X-ray images can play an essential role in mass screening for pulmonary tuberculosis. The introduction of deep convolutional neural network (CNN) models and open access publicly available large datasets made the wide spread application of computer vision algorithms. CNNs allow important image features to be learned automatically from the large training data, but acquiring annotated medical image datasets like ImageNet is a very challenging task [9,10,11]. X-ray imaging technique is a popular and a very low-cost modality, which can provide plenty of data to train machine learning models. Therefore, X-ray images are becoming popular in detecting lung abnormalities using deep CNN models.
Recently, several studies employed deep CNN models to detect lung abnormalities (i.e., pneumonia, lung cancer, tuberculosis) by analyzing CXR images [12,13,14,15]. Deep CNN models were extensively used to detect the novel coronavirus disease from CXR images[15,16,17,18]. Ismael and Sengur in [16] used a comparatively smaller dataset of CXR images and confirmed that deep learning had the potential for coronavirus disease 2019 (COVID-19) detection using CXR images. Features extracted using the ResNet50 model were classified using the support vector model (SVM) classifier with the linear kernel to produce an accuracy of 94.7%. Tahir et al. [12] proposed a framework for classifying coronavirus families with more than 90% sensitivity by utilizing multiple pre-trained CNN models. To differentiate viral pneumonia, COVID-19, and healthy patients, Chowdhury et al. [14] reported a deep CNN model for COVID-19 detection from CXR images while the different layers of the CNN model were used to identify the signature of viral pneumonia and COVID pneumonia in the X-ray images. Another study proposed a unique CNN model called PulDi-COVID for detecting nine different diseases, including COVID-19, using chest X-ray images and the SSE algorithm [19]. The test results showed that PulDi-COVID had high accuracy for identifying COVID-19 specifically with 99.70% accuracy, 98.68% precision, 98.67% recall, 98.67% F1 score, a low zero-one loss of 12 chest X-ray images, 99.24% AUC-ROC score, and a low error rate of 1.33%. A collection of recent literature on the use of X-ray, CT, and multimodal imaging for COVID-19 diagnosis was reviewed and classified based on the use of pre-trained and custom models by Yogesh and Patnaik in [20]. The authors also discussed the challenges of using deep learning for COVID-19 diagnostic systems and outlined areas for future research to improve the accuracy and reliability of COVID-19 detection. Another study proposed a unique CNN model called PulDi-COVID for detecting nine different diseases, including COVID-19, using chest X-ray images, and the Searchable symmetric encryption (SSE) algorithm [19]. The test results showed that PulDi-COVID had high accuracy for identifying COVID-19 specifically with 99.70% accuracy, 98.68% precision, 98.67% recall, 98.67% F1 score, a low zero-one loss of 12 chest X-ray images, 99.24% AUC-ROC score, and a low error rate of 1.33%. A collection of recent literature on the use of X-ray, CT, and multimodal imaging for COVID-19 diagnosis was reviewed and classified based on the use of pre-trained and custom models [20]. The authors also discussed the challenges of using deep learning for COVID-19 diagnostic systems and outlined areas for future research to improve the accuracy and reliability of COVID-19 detection. Ieracitano et al. [21] introduced a deep learning framework that incorporates fuzzy logic to distinguish between COVID-19 pneumonia and non-COVID-19 interstitial pneumonias based on chest X-ray (CXR) images. They utilized CXR images and fuzzy images generated through a formal fuzzy edge detection method as inputs to their developed CovNNet model, allowing for automatic extraction of the most crucial features. The experimental findings demonstrated that by combining CXR and fuzzy features, the classification performance significantly improved, reaching an accuracy rate of up to 81%.
Several research groups applied standard machine learning algorithms to stratify TB and healthy or other non-TB lung infections using CXR images [22,23,24,25,26,27]. Different groups have proposed deep CNN models [28,29,30,31,32,33] by pruning the networks to detect tuberculosis. TB patients were identified with an accuracy of 82.09% using a deep CNN model by Hooda et al. [28]. In [30], a CAD model was proposed for the detection of TB patients from the chest X-ray images with an accuracy of 88.76% utilizing important patterns in the lung images. Pasa et al. [31] showed a deep neural network for tuberculosis detection with an accuracy of 86.82%. They also mentioned a technique for interactively visualizing tuberculosis instances. In another work utilizing the ensemble of CNN models, Hernandez et al. [33] automatically classified TB patients from CXR images with an accuracy of 86%. Pre-trained CNN models, which were trained on the ImageNet dataset, were utilized by Lopes et al. [34] to stratify the TB and non-TB patients using CXR images. A simplified pre-trained CNN model for TB detection with and without image augmentation has been developed by Ahsan et al. [35] with an accuracy of 81.25% and 80%, respectively. Again, pre-trained CNN models were reported to show an accuracy of 94.89% in TB detection by Yadav et al. [36]. Abbas et al. [37] suggested a class decomposition strategy-based CNN architecture to enhance the performance of pre-trained models. It is worth noting here that TB culture test images were used for training the pre-trained CNN models. Chang et al. [38] achieved 98% sensitivity with 99% precision by applying the transfer learning technique to TB culture images. TB culture image-based classification needs specific samples from the patients, which makes it less reliable than classification from readily available chest X-rays. In our previous work [39], we presented a transfer learning approach utilizing deep Convolutional Neural Networks (CNNs) to automatically detect tuberculosis (TB) from chest radiographs. The researchers evaluated the performance of nine different CNN models in classifying TB and normal chest X-ray (CXR) images. Among these models, ChexNet demonstrated superior performance for datasets with the lung segmented CXR images. The results revealed high classification accuracy, precision, and recall for TB detection. Specifically, without segmentation, the accuracy, precision, and recall were found to be 96.47%, 96.62%, and 96.47%, respectively, while with segmentation, they increased to 98.6%, 98.57%, and 98.56%, respectively.
The drug-resistant TB strains are particularly concerning in the diagnosis and treatment of TB. There are currently around 20 medicines in use to treat tuberculosis. The five most often used medications, usually known as first-line treatments, are typically given to TB patients who did not get TB treatment before. Rifampin (RIF), isoniazid (INH), ethambutol (EMB), pyrazinamide (PZA), and streptomycin (SM) are the first-line drugs [40]. To avoid getting resistance to a single treatment, it is critical to take multiple TB drugs at the same time. Patients need to be very careful about the treatment plan for several months without missing a single dose to avoid drug resistance. Medicines for drug resistance tuberculosis, which are so-called second-line medications, have negative impacts as well as very expensive. These reserve drugs are grouped according to their experience of usage and efficiency. If the TB bacterium that causes the infection responds to all medicines, the patient is drug-susceptible or drug-sensitive. If a patient’s TB becomes drug-resistive, at least one of the primary medications will not affect the TB bacteria, either through poor treatment or transmitted by the infected patient. Multi-drug resistant TB (MDR-TB) and extreme drug-resistant TB (XDR-TB) are the two main kinds of drug resistance. MDR-TB is characterized as resistance to at least one of the most effective first-line TB medications, isoniazid or rifampicin. These two are the most common TB drug resistance types while further classifications are occasionally used relying on the number of medications to which the Mycobacterium tuberculosis bacteria stop responding such as resistance to specific drugs and resistance to the majority of currently available drugs. XDR TB is an extensively rare type of MDR TB, which is resistant to rifampin, fluoroquinolone, or any isoniazid drugs [41]. They are also resistant to any of the second-line injectable drugs (i.e., kanamycin, amikacin, or capreomycin). Patients have very fewer and less effective drug options due to the resistance of the XDR TB to the most potent TB drugs. Human immunodeficiency virus (HIV)-infected or other similar condition patient who has weak immune system should be very careful about XDR TB. Once infected such a patient can easily develop TB and also have a high risk of death after developing the TB.
MDR-TB is challenging to detect and requires additional time and cost for patient treatment (sometimes more than 2 years). MDR-TB affects 3.3% of new TB patients, as well as 20% of previously treated patients [42]. One of the most difficult aspects of treating MDR-TB is detecting drug resistance in suspected patients on their first hospital visit. Drug resistance status is determined by a drug susceptibility test often done on sputum samples. A well-equipped laboratory is necessary to acquire such a report in 4 to 6 weeks [43]. This investigation time can be considerably shortened to detect MDR-TB by the recent invention of the Xpert Mycobacterium tuberculosis/rifampicin (MTB/RIF) [44]. This is a real-time polymerase chain reaction test to identify the genetic changes that happened to the MTB genome related to rifampicin (RIF) drug resistance. But the sputum sample collection is still required for the test, which is very hard to collect, particularly from children. As a result, finding MDR-TB remains a difficulty, and because of its broad availability, the traditional chest X-ray (CXR) remains an important tool in the surveillance, diagnosis, and screening of MDR-TB. There is evidence that computed tomography (CT) images can be used to distinguish MDR TB and drug-sensitive TB. For instance, Yeom et al. [45] showed a substantial association between primary MDR-TB patients and multiple bilateral abnormalities in the lung CT images. Stefan et al. [44] reported multiple cavities and bilateral consolidations in the chest CT slices, which help in the discrimination analysis of MDR-TB patients. Chen et al. [46] used positron emission tomography and computed tomography (PET-CT) imaging to relate the abnormalities in images with the MDR TB patients while studying the changes in the lung abnormalities in a cohort of 28 MDR TB patients under second-line TB treatment for 2 years and then monitored for another 6 months using CT alone. Traditional sputum microbiology is less sensitive than several radiologic markers in detecting successful vs unsuccessful TB patients under treatment. Cha et al. [47] explained the radiological results of XDR-TB and compare them to those of MDR-TB and drug-sensitive TB among non-AIDS patients. Drug-sensitive TB was represented by the presence of several nodules, bronchial, and cavities dilatation in CT images of young individuals, whereas no significant difference in the image of the patients with MDR-TB and XDR-TB was observed. These findings were verified by Kim et al. [48], who detected apparent cavities in CT images for the patients with MDR-TB. This is also supported by the findings of Chung et al. [49]. On the other hand, Lee et al. [50] later determined that XDR-TB had more widespread consolidation and a tree-in-bud presence in CT images in comparison to MDR-TB. Very little effort has been made to automatically distinguish drug-resistant and drug-sensitive TB using CXR images. A significant relationship between the treatment resistance status of TB patients and computerized features of radiological imaging was identified by Kovalev et al. [51] in a pilot study. By combining CXR and CT features, the authors attained an accuracy of more than 75% in drug-resistance TB detection [52]. However, the CXR features alone had a substantially low performance. Stefan et al. [44] reported that it is possible to computationally extract relevant information from the chest X-ray images related to the drug-resistant TB infection. They have used the CXR images from the database of the Republic of Belarus where MDR/XDR-TB and HIV/TB are dominant. The database also incorporates the laboratory values and clinical biomarkers along with CXR images from either diagnosed or suspected MDR-TB patients. Out of 135 investigated cases by Stefan et al. [44], 45% (61) were sensitive while 54% (74) were MDR. As radiological images can provide details that can help in distinguishing the various drug-resistant TB categories, machine learning networks can be used to detect them and make decisions. It has been found from previous works of the authors and other recent work that novel machine learning networks along with pre-processing techniques can accurately detect other pulmonary abnormalities [13, 53, 54]. The above studies motivated this study to use a machine learning framework in classifying TB and healthy patients using chest X-ray images and further classify the TB patients into the different drug-resistant TB groups to help in early disease detection and treatment. The key contributions of this work are highlighted below:
-
The largest TB benchmark dataset, namely, QU-MLG-TB, has been created using 40,000 CXR images along with their ground truth lung masks. Of the TB patients’ CXR images, 10,881 normal (healthy), 24,119 non-TB (other lung infections), and 5000 are present in the dataset. This is the largest TB dataset which is collected from multiple open access and restricted access databases.
-
A novel framework, TB-CXR-Net, for TB detection using this largest dataset was proposed. This is a benchmark performance on a benchmark dataset with the highest accuracy ever achieved in the diagnosis and assessment of TB disease using CXR alone.
-
The largest drug-resistant TB dataset as a subset of the QU-MLG-TB dataset will be released to develop a machine learning model for drug-resistant TB detection and stratification.
-
A state-of-the-art machine learning stacking model is proposed to detect and stratify drug-resistant TB from chest X-ray images alone with state-of-the-art performance.
-
Score-CAM-based visualization technique to see how the best performing model decides to classify the image.
The paper is divided into five subsequent sections. The “Methodology” section summarizes the methodology used in the paper with the details of the datasets and pre-processing steps. The “Experiments” section provides the experimental details, while the “Results and Discussion” section describes the results and discussion of the TB classification and drug-resistant TB stratification. Finally, the article is concluded in the “Conclusion” section.
Methodology
The objective of this research is first to classify the TB patients among healthy control and non-TB other lung infections and then stratify the TB patients into drug-sensitive and drug-resistive TB.
In the first stage, a novel framework was developed to classify normal, non-TB (other lung infections), and TB patients (Fig. 1A) using a convolutional neural network with a non-linear neuron-based multi-layer perceptron (MLP) classifier [55]. In the second stage, the chest X-ray images of the TB patients are applied as input to a CheXNet-based CNN encoder to extract CXR image features, and then the dimensionality of the extracted features was reduced using the principal component analysis (PCA). Finally, different machine learning classifiers and stacking approaches were investigated to find the best performing model to classify the TB chest X-rays into binary (drug-resistant and drug-sensitive, i.e., cases that are sensitive to all the TB drugs (Fig. 1B)) and 3-class problems (MDR, XDR, and sensitive-TB (Fig. 1B)). The overall methodology of the proposed system is shown in Fig. 1.
Dataset Description
The study considered only posterior-to-anterior or anterior-to-posterior view of the chest X-ray images, as this view is widely used by radiologists. QU-MLG-TB is the largest TB benchmark dataset and consists of 40,000 chest X-ray (CXR) images along with their corresponding lung masks. Details of the full dataset are shown in Table 1. There are two categories in the dataset:
TB Classification
This is one of the largest datasets for TB which consist of 10,881 normal (healthy), 24,119 non-TB (other lung infections), and 5000 TB patients’ CXR images that are present in the dataset. This dataset was built using a variety of publicly available and restricted access datasets and repositories [39, 53]. Therefore, the dataset has a wide range of resolution and format and was collected using different equipment. In the pre-processing phase, the authors identified and discarded the images with extremely low-quality, over-exposed, and duplicate images to ensure a good quality dataset for this study.
RSNA CXR Dataset (Non-COVID Infections and Normal CXR)
The RSNA pneumonia detection challenge dataset [56] is made up of 26,684 chest X-ray images, where 8851 images are normal, 11,821 are abnormal, and 6012 are images of lung opacity. The images are in DICOM format. In this study, we used 8851 normal images and 6012 images of lung opacity as the non-COVID class.
PadChest Dataset
The PadChest dataset [57] is made up of more than 160,000 X-ray images from 67,000 patients that were collected and reported by radiologists at Hospital San Juan (Spain) from 2009 to 2017. In this study, we used 4000 normal and 4000 pneumonia/infiltrate (non-COVID-19) cases from the PadChest dataset.
NLM Dataset
The National Library of Medicine (NLM) in the USA has made two datasets of lung X-ray images publicly available: the Montgomery and Shenzhen datasets [58]. The Montgomery County (MC) and the Shenzhen, China (CHN) databases consist of 138 and 667 posterior-anterior (PA) chest X-ray images, respectively. The resolution of the images in the MC database is either 4020 × 4892 or 4892 × 4020 pixels, while the resolution of the images in the CHN database is variable, but around 3000 × 3000 pixels. In the MC database, out of the 138 chest X-ray images, 58 were taken from different TB patients, and 80 were from normal subjects. In the CHN database, out of 662 chest X-ray images, 336 were taken from different TB patients, and 324 were from normal subjects. Therefore, in this NLM database, there are 406 normal and 394 TB-infected X-ray images.
Belarus Dataset
The Belarus dataset [59] was gathered for a study on drug resistance led by the National Institute of Allergy and Infectious Diseases, Ministry of Health, Republic of Belarus. The dataset includes 306 chest X-ray images from 169 patients. The images were taken using the Kodak Point-of-Care 260 system and have a resolution of 2248 × 2248 pixels. All the images in this database are from individuals infected with TB.
NIAID TB Dataset
The NIAID TB portal program dataset [60] includes around 3037 chest X-ray images that are positive for TB from approximately 3087 cases. The images were collected from seven different countries and are in Portable Network Graphics (PNG) format.
Drug-Resistant TB Classification
A subset of the QU-MLG-TB dataset, where 3037 CXR images out of 5000 TB images labeled as drug-resistant/sensitive TB, was used for the drug-resistant TB classification. Among these 3037 CXR images, 626, 1672, and 739 images are sensitive, MDR, TB, and XDR TB, respectively. Figure 2 (last row) shows the sample CXR images of the drug-resistant/sensitive TB. Figure 2 shows the sample images for healthy, non-TB other lung infections, and TB images with high interclass variations and varied quality, signal-to-noise ratio (SNR) levels, and resolution. Figure 2 (last row) shows the sample CXR images of the drug-resistant/sensitive TB. Figure 2 (last row) shows the sample CXR images of the drug-resistant/sensitive TB.
Preprocessing
This section describes different pre-processing steps used in this study, such as image enhancement techniques, technical details in the model development for the lung segmentation and classification including feature extraction, feature reduction using principal component analysis (PCA), and finally stacking machine learning-based classification.
Gamma Correction
The image enhancement technique is to is to highlight important information in an image while reducing or eliminating irrelevant details, thereby enhancing decision-making performance. In this study, the authors employed the Gamma correction technique, which has previously demonstrated improved classification performance on chest X-ray (CXR) images in the works of the same authors [39, 53]. While linear operations such as addition, subtraction, and scalar multiplication are commonly used for pixel normalization in image processing, Gamma correction involves applying a non-linear operation to enhance the pixels of the image. Gamma correction is typically denoted by the following expression:
where the non-negative pixel values are raised to the power of γ and gamma value can be greater or smaller than 1 and multiplied by the constant A.
Lung Segmentation Model Development
It is very important to localize the region of interest for the machine learning networks, i.e., the lungs in the chest X-ray images. In our previous work for CXR lung segmentation [61], a detailed investigation was done on three segmentation architectures, Feature Pyramid Networks (FPN) [62], U-Net++ [63], and U-Net [64] with various encoder backbones. FPN [62] segmentation network with DenseNet121 [65] encoder as a backbone outperformed other conventional segmentation networks [61]. Using the FPN network with DenseNet121 backbone, the lung area is segmented very accurately which was verified by the experienced radiologists in the previous work. The model trained in [61] was used to create lung segmentation for this work. Figure 3 shows the sample chest X-ray images and their corresponding lung masks.
TB Classification Model and Drug-Resistant TB Stratification Model
Two experimental frameworks have been proposed in this study to classify TB patients into healthy and non-TB other lung infections and then stratify drug-resistive TB patients among the TB patients.
TB Classification Model
In this study, a novel deep learning network using a pre-trained CNN (ChexNet) encoder and non-linear neuron-based MLP classifier (Self-MLP)—details provided below, is proposed for TB classification (Fig. 4).
Feature Extractor
An encoder of the pre-trained CNN model, ChexNet, was used to extract important features from the segmented chest X-rays. It should be worth mentioning here that CheXNet is a variant of DenseNet (DenseNet121) which was trained on a large chest X-ray dataset and the pre-trained model is available publicly. CheXNet performed exceptionally well in the CXR image classification task for COVID-19 as shown in our previous work [39]. In the DenseNet architecture, in the Dense block, each layer is connected to every other layer. Every Dense block has a feature map of the same size, and the features are reused within the network. Such Dense connections connecting the DenseNet layers expedite the flow of information throughout the network. The useful features of the CXR images from the CheXNet were extracted from the last layer of the encoder, ‘AvgPool.’
The DenseNet encoder has three dense blocks that each has an equal number of layers. Before entering the first dense block, a convolution with 16 output channels is performed on the input images. For convolutional layers with kernel size 3 × 3, each side of the inputs is zero-padded by one pixel to keep the feature-map size fixed. Moreover, the encoder part has 1 × 1convolution followed by 2 × 2 average pooling as transition layers between two contiguous dense blocks. At the end of the last dense block, a global max pooling is performed and then a Self-MLP classifier is attached. The feature-map sizes in the three dense blocks are 32 × 32, 16 × 16, and 8 × 8, respectively.
Principal Component Analysis-Based Feature Reduction
Principal component analysis (PCA) is used to reduce the dimensionality of the feature space extracted from the ChexNet encoder. PCA projects high-dimensional data into a new lower-dimensional representation with as minimal reconstruction error as feasible. Because all the fundamental components in the reduced set are orthogonal to one another, there is no redundant data. PCA was calculated with the use of whitening, which can improve accuracy by forcing data to meet certain assumptions.
Self-MLP
To overcome the linear nature of CNN, the Operational Neural Network (ONN)-based model was recently presented in [55]. ONN is a heterogeneous network that learns complicated patterns of any signal using a fixed set of non-linear operators and has demonstrated promising results in numerous applications such as image denoising and image restoration [66,67,68,69]. Self-organized Operational Neural Networks (Self-ONN) is a new variant of ONN. Instead of a fixed collection of operator libraries, Self-ONN learns the best set of operators throughout the training process. This results in a more robust model that can handle a wider range of scenarios and generalizes effectively in real-world scenarios. During the training phase, operational layers determine the best set of operators, which can be a combination of any conventional functions or unknown functions. The output \({x}_{k}^{l}\) at \(k\) th neuron of \({l}\) th the layer of any ONN can be illustrated as follows in Eqs. (2) and (3):
where \({b}_{k}^{l}\) and \({w}_{ki}^{l}\) denote the biases and weights corresponding with that neuron and layer, \({y}_{i}^{l-1}\) represent the previous layer’s input, \({N}_{l-1}\) stands for kernel size of that layer, and \({\Psi }_{ki}^{l}\) corresponds to the nodal operator of the neuron and layer. If \({\Psi }_{ki}^{l}\) is linear then the equation simply corresponds to conventional CNN. In ONN, the composite nodal operator \(\Psi\) can be constructed using a set of standard functions as follows:
where \(\mathbf{w}\) represents the q-dimensional array of parameters that are composed of internal parameters and weights of the individual functions. Instead of a fixed set of operators, the composite nodal operator \(\Psi\) can be constructed using a Taylor series approximation. The Taylor series approximation of a function \(f(x),\) near point, \(x=a\) is expressed by the following equation:
Equation (4) can be used to construct the nodal operator as follows:
where \({w}_{q}=\frac{{f}^{(n)}(a)}{q!}\) is the \(q\) th parameter of the \(q\) th-order polynomial. In Self-ONN, tangent hyperbolic (tanh) has been used as an activation function that is bounded at the range [− 1, 1]. So, for tanh, \(a\) is equal to zero in Eq. (5).
Figure 4 illustrates the CheXNet-Self-MLP-based TB classifier that uses Self-MLP as a classifier after the CheXNet-based encoder. MLP layers can be implemented using convolutional layers by using kernels of the same size as the input. Thus, a single sliding window of the convolutional kernel will cover the full signal, retaining the fully connected nature of MLPs. Similarly, 1D operational layers can be used to implement Self-MLP layers, which were used in the implementation of this study.
Drug-Resistant TB Stratification Model
ML Classifier
To identify the patients with TB drug resistance, the CheXNet encoder was used to extract features as mentioned earlier, PCA was used to reduce the features obtained from the encoder and then eight machine learning classifiers, including Support Vector Machine (SVM) [70], K-nearest neighbor (KNN) [71], XGBoost [72], Random Forest [73], Adaboost [74], linear discriminant analysis (LDA) [75], Gradient boosting [76], and Logistic regression [77], were used to classify a subset of TB patients into sensitive and drug-resistive TB (binary class problem) and XDR, MDR, and sensitive TB (3-class problem).
Stacking Model
The three best performing classifiers were chosen as base learner models (M1, M2, M3) in the stacking architecture, and a meta learner classifier (Mf) was trained in the second phase, resulting in separate performance matrices based on the final prediction. Consider a single dataset A is consisting of input vectors (\({x}_{i}\)) and their classification score (\({y}_{i}\)). At first, a set of base-level ML classifiers \({M}_{1},\dots \dots ,{M}_{p }\) is trained on the dataset and the estimation of these base learners is applied to train the meta-level classifier\({M}_{f }\mathrm{}\), [78,79,80] which is illustrated in Fig. 5.
Model Interpretability
Saliency map or class activation map techniques make the deep learning model interpretable. It is become very important to see why the CNN model works and to know the underlying reason in the decision-making process. This helps to make the model trustworthy as the reason for classification becomes evident to humans. SmoothGrad [81], Grad-CAM [82], Grad-CAM++ [83], and Score-CAM [84] are the common visualization techniques. However, Score-CAM was used in this study as it outperformed other techniques in the recently reported medical image classification problems. The heat map created by the Score-CAM technique will show the regions of the images where the model is learning most. This visualization process of CNN allows its user to have trust in the model decision if it is seen that the model is learning from the relevant area of the image rather than just using the model as back-box without any clue where is taking the decision from.
Experiments
Different experiments conducted in this study are listed in this section.
TB Classification
A novel model, CheXNet-Self-ONN, using a pre-trained CheXNet encoder with a Self-MLP classifier was trained, tested, and validated for the classification of TB, non-TB (other lung infections), and healthy CXR images. Four other state-of-the-art pre-trained CNN models were used as feature extractors (encoder) to compare the performance with the CheXNet-based encoder. The image dataset was partitioned into 80–20% train-test sets and 20% of the training set was kept aside for validation. This process is repeated 5 times to get five-fold cross-validation results.
However, the number of images in the training set per fold for different classes is not equal, and therefore, the training images for TB and healthy classes were augmented to balance the training dataset. Three different image augmentation techniques such as translation, rotation, and scaling [85] were utilized to balance the training data classes and expand the training set. The healthy class was augmented one time (translation by 10% from top and left) only while the TB class was augmented four times (rotation by 5 and 10°, translation by 10% from top and left, and translation by 10% from top and right). The details of the train, validation, and test sets for the TB classification task are shown in Table 2.
Drug-Resistant TB Stratification
This stratification framework includes a CheXNet encoder, PCA for dimensionality reduction, and classification using the stacking ML model. CheXNet pre-trained model was used to extract spatial features from the lung segmented CXR images and performed PCA to reduce the dimensionality. Then eight state-of-the-art ML classifiers and a stacking ML model were investigated to predict different drug-resistant patients using the PCA reduced features. This experiment was done also using five-fold stratified cross-validation, where 80% of drug-resistive/sensitive images in each class were utilized for training and 20% of images per class were used as test set and validation set (20% of train set) were used to avoid over-fitting. Rotation, translation, and scaling were utilized as image augmentation techniques to balance the training data classes and expand the training set to the make the number of images available in the training set equal to the number of images present in the class of maximum image (i.e., 1337 for MDR class in this example). Image rotation rotates the original image either clockwise or counterclockwise (5° to 10°). The scaling process expands or shrinks the image, with image magnifications ranging from 2.5 to 10% in this investigation. Image translation was performed with 5–10% horizontal or vertical translation. The details of the train, validation and test set used in this study are reported in Table 2. To report the result, a weighted average of the five-folds was obtained.
Training Parameters
All classification models were instigated using Python 3.7 and PyTorch library, a 64-GB RAM, and a 16-GB NVIDIA GeForce GTX 1080 GPU using an Intel® Xeon® E5-2697v4 processor running at 2.30 GHz. Training parameters and stopping criteria were the same for all classification models and the models were trained with 15 backpropagation epochs. The results of five-fold test sets were accumulated to generate the receiver operating characteristic (ROC) curve and confusion matrix and to calculate overall accuracy and weighted evaluation metrics. Details of the training parameters are also summarized in Table 3.
Performance Matrix for TB and Drug-Resistant TB Classification
The performance of different classifiers was evaluated using ROC curves along with the area under the curve (AUC) as well as precision, sensitivity, specificity, accuracy, and F1 score. Since five-fold cross-validation was used in this study, the reported results are on the entire data (five test fold-concatenated). Because different classes had variable numbers of instances, weighted metrics per class and overall accuracy were reported. As a metric for performance comparison, the area under the curve (AUC) was considered. The mathematical expressions of five evaluation metrics such as weighted sensitivity, precision, specificity, F1 score, and overall accuracy are reported below:
where class_i are the healthy, non-TB, and TB or drug-resistant TB and sensitive TB.
Here, false positive, false negative, true positive, and true negative are represented as FP, FN, TP, and TN, respectively. TP represents how many of the positive class was correctly identified, and TN represents how many of the negative class was correctly identified. FP represents how many of the negative class was incorrectly identified as positive and FN represents how many of the positive class was incorrectly identified as negative.
Results and Discussion
This section showcases the results from the different experiments conducted in the study along with the discussions about the findings.
TB Classification
The comparative performance of the proposed CheXNet-Self-MLP model and various pre-trained CNN models for the 3-class classification are shown in Table 4 below. According to Table 4, all the evaluated pre-trained models perform very well when it comes to classifying TB, non-TB, and healthy images in this three-class problem. The proposed model outperformed all the various pre-trained CNN Encoder-Self-MLP models in TB prediction. Figure 6 illustrates that the performance of the models is comparable and the ROC curves showed similar performances from all networks. Although different pre-trained CNN models show good performance in 3-class classification, CheXNet-Self-MLP displayed the best performance in classifying healthy, non-TB, and TB chest X-ray images with accuracy, recall, and F1 score of 93.2%, 93.38%, and 94.58%, respectively. The second best performing model was DenseNet201, which showed an accuracy, recall, and F1 score of 92.45%, 92.47%, and 93.03%, respectively.
Figure 6(A) clearly shows the ROC curves for various CNNs and the proposed CheXNet-Self-MLP model, where the proposed model generated an AUC of 0.9887 and outperformed various state-of-the-art CNN models. It is also evident that the pre-trained CNN models performed well for the 3-class classification. The confusion matrix for the best performing model, CheXNet-Self-MLP, is shown in Fig. 6(B). It can be noticed that 4628 out of 5000 TB images were correctly classified by the model. With this exceptional performance of the computer-aided classifier, the proposed model can help the radiologists significantly in screening TB patients using only chest X-ray images.
Drug-Resistant TB Classification
Two different approaches were investigated for drug-resistant TB classification using CXR images. Firstly, PCA-reduced CXR features were used for binary classification (sensitive vs drug-resistant TB). Secondly, three-class classifications (sensitive, MDR, and XDR TB) were investigated using CXR images. For both investigations, eight different machine learning classifiers were analyzed, and a novel stacking model was constructed to compare with the eight different machine learning classifiers.
For binary classification, the Gradient boosting classifier was the best performing classifier. It achieves the accuracy, precision, sensitivity, and F1 scores of 86.43%, 85.7%, 86.43%, and 85.05%, respectively. Then the stacking model was built using the top three performing classifiers (i.e., Gradient boosting, Adaboost, and Logistic regression). The stacking model produces better performance with the accuracy, precision, sensitivity, and F1 scores of 87.85%, 87.46%, 87.84%, and 86.65%, respectively, which is ~ 2% better than the best performing model, the Gradient boosting classifier.
Similarly, for three-class classification, the Gradient boosting classifier was the best performing classifier. It achieves the accuracy, precision, sensitivity, and F1 scores of 77.35%, 77.27%, 77.35%, and 74.16%, respectively. The stacking model was built using the top three classifiers (i.e., Gradient boosting, Adaboost, and XGboost). The stacking model also produces better performance with the accuracy, precision, sensitivity, and F1 scores of 79.59%, 78.78%, 79.59%, and 78.33%, respectively, which is ~2% better than the best performing Gradient boosting classifier. Table 5 shows the performance of eight different state-of-the-art machine learning classifiers with stacking models for binary and three-class classification.
Figure 7(A) displays the ROC curves for binary classification (drug-resistant vs sensitive) using the proposed stacking model and different ML classifiers, where the stacking model provided 0.912 AUC and outperforms all other ML classifiers. It is also shown that the Gradient boosting classifier performed the second-best with an AUC of 0.904. Figure 7(B) also clearly shows the ROC curves for three-class (MDR, XDR, and sensitive TB) classification using eight ML classifiers and the new stacking model. It is also noticed that the stacking ML model produced the best performance compared to other ML models with an AUC of 0.808. The stacking ML model showed a 2% improvement in AUC compared to the best performing classifier (Gradient boosting classifier).
Figure 8 shows the confusion matrix for outperforming the stacking model in classifying drug-sensitive TB and drug-resistive TB using chest X-ray images for 2-class and 3-class classification. It can be noticed that 2116 out of 2411 drug-resistant TB images were correctly classified by the stacking model for binary classification whereas 1600 out of 1672 MDR TB images were correctly classified by the stacking model for three-class classification. It is evident from Fig. 8(A) that the proposed model can stratify the drug-resistive TB and drug-sensitive TB with very high accuracy, which is the state-of-the-art performance to the best of the authors’ knowledge compared to all work reported in the literature (Table 6).
The proposed model is also trained on a very large and robust dataset [86], which also confirms its reliability and generalizability. It is also reported in the literature that there is a distinct pattern of drug-resistant TB which makes it possible to be identified from chest X-rays. The non-linearity provided by the Self-MLP has helped in identifying this pattern. The authors also wanted to investigate if it was possible to distinguish MDR and XDR TB patients from chest X-rays to facilitate early drug-resistant TB detection and avoid spreading the infection. As seen in Fig. 8(B), the proposed model can detect MDR-TB reliably while many misclassification happens between MDR and XDR TB groups. It is understood that additional clinical information is required to classify them more accurately, which is a future direction of work authors are planning.
Model Interpretability
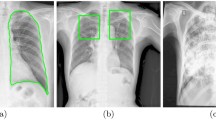
As mentioned earlier, this study used the Score-CAM-based visualization technique to see how the best performing model decides to classify the image, which is the image it considers for taking the decision. Since the model input is the segmented lung images and the infections investigated in this study are confined to the lung only, the model is constrained to learn from the lung only. However, the model still can learn from the area of infection or non-infectious area of the lung, which can be verified by the Score-CAM technique. Figure 9(A)–(C) show the raw chest X-ray images with lesion and consolidation (first row) and Score-CAM visualization of the best performing model (bottom row) for (A) healthy, (B) non-TB, and (C) TB subjects. Figure 9(A) shows that the model is learning from the entire image and there is no specific area in the lung from where the model is taking decisions.
In Fig. 9(B), it is visible that Score-CAM-based heat maps were learned mostly from the same area where both images are showing the abnormality (highlighted the abnormal region by a red arrow). Both the CXR images are non-TB lung infections with lung opacity in both of the lungs. The Score-CAM images highlight those areas as the most important areas for taking decisions. Figure 9(C) shows the lymph nodes in the lower right lung of the first image and upper right lung of the second image in TB patients. Score-CAM-based heat maps showed the same area as the most contributing area in the decision of CNN. Thus, it can be concluded that the model decided on the abnormal region of CXR images. Moreover, the patterns of abnormal areas in the lungs for non-TB and TB are quite different where non-TB CXR images are showing abnormalities in both lungs’ middle and lower parts but TB images are mostly showing the consolidation, cavitation, and other abnormalities in one of the lungs.
The study also used the Score-CAM visualization technique for the second experiment to see how the best performing model learns from the lung images and which area of the lung is used for making the decision. Figure 10 shows the raw chest X-ray images of MDR-TB, XDR-TB, and sensitive-TB subjects with lesion and consolidation (first row: A–C) and Score-CAM visualization for the best performing model. It is apparent that Score-CAM-based heat maps show the highest important area in CXR images corresponding to the area of abnormality as shown by red arrows.
It is also interesting to see the Score-CAM visualization for misclassified CXR TB images. Figure 11 shows the CXR images for TB patients misclassified into other classes where the first two CXR TB images (from left) are misclassified as normal and the last two are misclassified as non-TB patients.
The first two cases of CXR images of TB patients misclassified as normal can be explained as the patients were in the initial stage of TB disease development and thus the effect of TB is not yet visible on the CXR images. The last two TB images misclassified as non-TB can be understood from the heat map where the pattern is different from most of the TB cases. In these cases, both the lungs are largely affected, while typically in TB images only the lung is affected (Fig. 9(C)). These misclassifications can be avoided by the use of multimodal techniques where CXR images can be used with clinical biomarkers to improve the performance of the model; this will be investigated in a prospective study in the future. In summary, the proposed work confirms that it is possible to detect drug-resistant TB from chest X-ray images and this work produces better performance compared to the ones reported in the literature (Table 5).
Conclusion
This work describes a novel stacking method to diagnose tuberculosis and drug-resistant tuberculosis from chest X-ray images. The entire framework is divided into two steps: firstly, TB-infected CXR images were identified among the TB, non-TB (other lung infections), and healthy CXR images, and secondly, the TB CXR images are further stratified into sensitive TB or drug-resistive TB images. It was found that the CheXNet encoder with Self-MLP outperforms compared to other pre-trained deep learning models with an accuracy of 93.2% using segmented lung CXR images. The paper also proposes a novel stacking approach to classify TB chest X-rays as drug-resistant and drug-sensitive TB with 80% accuracy which is the best-reported result in the literature. The work could be further improved with the help of a larger labelled dataset and multimodal approach using clinical information from Electronic Health Record (EHR) data can also help in improving the performance. The proposed novel TB detection and drug-resistant TB stratification framework can significantly help in the timely detection of this killer disease and can help in taking proactive action to avoid the spreading of disease and infection. The proposed rapid diagnostic tool with such high performance can be a very valuable tool to save a significant number of people from death due to improper and delayed diagnoses every year.
Data Availability
The dataset used in this study can be made available with a reasonable request to the corresponding author.
References
Fukunaga R, Glaziou P, Harris JB, Date A, Floyd K, Kasaeva T. Epidemiology of tuberculosis and progress toward meeting global targets—worldwide, 2019. Morb Mortal Wkly Rep. 2021;70(12):427.
Sharma SK, Mohan A. Tuberculosis: From an incurable scourge to a curable disease-journey over a millennium. Ind J Med Res. 2013;137(3):455.
Silverman C. An appraisal of the contribution of mass radiography in the discovery of pulmonary tuberculosis. Am Rev Tuberc. 1949;60(4):466–82.
van’t Hoog AH, Meme HK, Laserson KF, Agaya JA, Muchiri BG, Githui WA, Odeny LO, Marston BJ, Borgdorff MW. Screening strategies for tuberculosis prevalence surveys: the value of chest radiography and symptoms. PloS one. 2012;7(7):e38691.
Brady AP. Error and discrepancy in radiology: inevitable or avoidable? Insights Into Imaging. 2017;8(1):171–82.
Degnan AJ, Ghobadi EH, Hardy P, Krupinski E, Scali EP, Stratchko L, et al. Perceptual and interpretive error in diagnostic radiology—causes and potential solutions. Acad Radiol. 2019;26(6):833–45.
Van Cleeff M, Kivihya-Ndugga L, Meme H, Odhiambo J, Klatser PJBID. The role and performance of chest X-ray for the diagnosis of tuberculosis: a cost-effectiveness analysis in Nairobi, Kenya. 2005;5(1):111.
Graham S, Das Gupta K, Hidvegi R, Hanson R, Kosiuk J, Al Zahrani K, et al. Chest radiograph abnormalities associated with tuberculosis: reproducibility and yield of active cases. 2002;6(2):137–42.
Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–98.
Greenspan H, Van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Transactions on Medical Imaging. 2016;35(5):1153–9.
Ravishankar H, Sudhakar P, Venkataramani R, Thiruvenkadam S, Annangi P, Babu N, et al. Understanding the mechanisms of deep transfer learning for medical images. In: Deep learning and data labeling for medical applications. Springer; 2016. p. 188–96.
Tahir AM, Qiblawey Y, Khandakar A, Rahman T, Khurshid U, Musharavati F, et al. Deep learning for reliable classification of COVID-19, MERS, and SARS from chest X-ray images. Cognit Comput. 2022;1–21.
Rahman T, Chowdhury ME, Khandakar A, Islam KR, Islam KF, Mahbub ZB, et al. Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray. Appl Sci. 2020;10(9):3233.
Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–76.
Qiblawey Y, Tahir A, Chowdhury ME, Khandakar A, Kiranyaz S, Rahman T, et al. Detection and severity classification of COVID-19 in CT images using deep learning. Diagnostics. 2021;11(5):893.
Ismael AM, Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst Appl. 2021;164:114054.
Ghaderzadeh M, Asadi F. Deep learning in the detection and diagnosis of COVID-19 using radiology modalities: a systematic review. J Healthc Eng. 2021;2021.
Aishwarya T, Ravi Kumar V. Machine learning and deep learning approaches to analyze and detect COVID-19: a review. SN comput sci. 2021;2(3):1–9.
Bhosale YH, Patnaik KS. PulDi-COVID: chronic obstructive pulmonary (lung) diseases with COVID-19 classification using ensemble deep convolutional neural network from chest X-ray images to minimize severity and mortality rates. Biomed Signal Process Control. 2023;81: 104445.
Bhosale YH, Patnaik KS. Application of deep learning techniques in diagnosis of COVID-19 (coronavirus): a systematic review. Neural Process Lett. 2022;1–53.
Ieracitano C, Mammone N, Versaci M, Varone G, Ali A-R, Armentano A, et al. A fuzzy-enhanced deep learning approach for early detection of COVID-19 pneumonia from portable chest X-ray images. Neurocomputing. 2022;481:202–15.
Das H, Nath A. An efficient detection of tuberculosis from chest X-rays. International Journal of Advance Research in Computer Science and Management Studies. 2015;3(5).
Melendez J, Sánchez CI, Philipsen RH, Maduskar P, Dawson R, Theron G, et al. An automated tuberculosis screening strategy combining X-ray-based computer-aided detection and clinical information. Sci Rep. 2016;6:25265.
Singh N, Hamde S. Tuberculosis detection using shape and texture features of chest X-rays. In: Innovations in electronics and communication engineering: proceedings of the 7th ICIECE 2018. Singapore: Springer; 2019. p. 43–50.
Van Ginneken B, Katsuragawa S, ter Haar Romeny BM, Doi K, Viergever MA. Automatic detection of abnormalities in chest radiographs using local texture analysis. IEEE Transa Med Imaging. 2002;21(2):139–49.
Jaeger S, Karargyris A, Antani S, Thoma G. Detecting tuberculosis in radiographs using combined lung masks. In 2012 Annual international conference of the IEEE engineering in medicine and biology society. 2012;2012:4978–81.
Melendez J, Sánchez CI, Philipsen RH, Maduskar P, van Ginneken B. Multiple-instance learning for computer-aided detection of tuberculosis. In Medical Imaging 2014: Computer-Aided Diagnosis. 2014;90351J.
Hooda R, Sofat S, Kaur S, Mittal A, Meriaudeau F. Deep-learning: A potential method for tuberculosis detection using chest radiography. In: 2017 IEEE international conference on signal and image processing applications (ICSIPA). IEEE; 2017. p. 497–502.
Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–82.
Evalgelista LGC, Guedes EB. Computer-aided tuberculosis detection from chest X-ray images with convolutional neural networks. In Anais do XV Encontro Nacional de Inteligência Artificial e Computacional. 2018;518–27.
Pasa F, Golkov V, Pfeiffer F, Cremers D, Pfeiffer D. Efficient deep network architectures for fast chest X-ray tuberculosis screening and visualization. Sci Rep. 2019;9(1):1–9.
Nguyen QH, Nguyen BP, Dao SD, Unnikrishnan B, Dhingra R, Ravichandran SR, et al. Deep learning models for tuberculosis detection from chest X-ray images. In 2019 26th International Conference on Telecommunications (ICT). 2019;381–6.
Hernández A, Panizo Á, Camacho D. An ensemble algorithm based on deep learning for tuberculosis classification. In: International Conference on Intelligent Data Engineering and Automated Learning, 20th International Conference. UK: Manchester; 2019. p. 145–54.
Lopes U, Valiati JF. Pre-trained convolutional neural networks as feature extractors for tuberculosis detection. Comput Biol Med. 2017;89:135–43.
Ahsan M, Gomes R, Denton A. Application of a Convolutional Neural Network using transfer learning for tuberculosis detection, in. IEEE International Conference on Electro Information Technology (EIT). 2019;2019:427–33.
Yadav O, Passi K, Jain CK. Using deep learning to classify X-ray images of potential tuberculosis patients, in. IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2018;2018:2368–75.
Abbas A, Abdelsamea MM, Gaber MM. Detrac: transfer learning of class decomposed medical images in convolutional neural networks. IEEE Access. 2020;8:74901–13.
Chang R-I, Chiu Y-H, Lin J-W. Two-stage classification of tuberculosis culture diagnosis using convolutional neural network with transfer learning. J Supercomput. 2020;1–16.
Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Kashem SBA, et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput Biol Med. 2021;132: 104319.
Verboven L, Callens S, Black J, Maartens G, Dooley KE, Potgieter S, Cartuyvels R, Laukens K, Warren RM, Van Rie A. A machine-learning based model for automated recommendation of individualized treatment of rifampicin-resistant tuberculosis. 2023.
Migliori GB, Tiberi S, Zumla A, Petersen E, Chakaya JM, Wejse C, et al. MDR/XDR-TB management of patients and contacts: challenges facing the new decade. The 2020 clinical update by the Global Tuberculosis Network. Int J Infect Dis. 2020;92:S15–25.
Chen M, Miramontes R, Kammerer J. Multidrug-resistant tuberculosis in the United States, 2011–2016: patient characteristics and risk factors. Int J Tuberc Lung Dis. 2020;24(1):92–9.
Parsons LM, Somoskövi Á, Gutierrez C, Lee E, Paramasivan C, A. l. Abimiku, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev. 2011;24(2):314–50.
Jaeger S, Juarez-Espinosa OH, Candemir S, Poostchi M, Yang F, Kim L, et al. Detecting drug-resistant tuberculosis in chest radiographs. Int J Comput Assist Radiology Surg. 2018;13(12):1915–25.
Yeom JA, Jeong YJ, Jeon D, Kim K-I, Kim CW, Park HK, et al. Imaging findings of primary multidrug-resistant tuberculosis: a comparison with findings of drug-sensitive tuberculosis. J Comput Assist Tomogr. 2009;33(6):956–60.
Chen RY, Dodd LE, Lee M, Paripati P, Hammoud DA, Mountz JM, et al. PET/CT imaging correlates with treatment outcome in patients with multidrug-resistant tuberculosis. Sci Transl Med. 2014;6(265):265ra166-265ra166.
Cha J, Lee HY, Lee KS, Koh W-J, Kwon OJ, Chin AY, et al. Radiological findings of extensively drug-resistant pulmonary tuberculosis in non-AIDS adults: comparisons with findings of multidrug-resistant and drug-sensitive tuberculosis. Korean J Radiol. 2009;10(3):207–16.
Kim H-C, Goo JM, Lee HJ, Park SH, Park CM, Kim TJ, et al. Multidrug-resistant tuberculosis versus drug-sensitive tuberculosis in human immunodeficiency virus-negative patients: computed tomography features. J Comput Assist Tomogr. 2004;28(3):366–71.
Chung MJ, Lee KS, Koh W-J, Kim TS, Kang EY, Kim SM, et al. Drug-sensitive tuberculosis, multidrug-resistant tuberculosis, and nontuberculous mycobacterial pulmonary disease in nonAIDS adults: comparisons of thin-section CT findings. Eur Radiol. 2006;16(9):1934–41.
Lee ES, Park CM, Goo JM, Yim J-J, Kim H-R, Lee HJ, et al. Computed tomography features of extensively drug-resistant pulmonary tuberculosis in non-HIV-infected patients. J Comput Assist Tomogr. 2010;34(4):559–63.
Kovalev V, Liauchuk V, Safonau I, Astrauko A, Skrahina A, Tarasau A. Is there any correlation between the drug resistance and structural features of radiological images of lung tuberculosis patients. In Computer Assisted Radiology -27th International Congress and Exhibition (CARS-2013). 2013;18–20.
Kovalev V, Liauchuk V, Kalinovsky A, Rosenthal A, Gabrielian A, Skrahina A, et al. Utilizing radiological images for predicting drug resistance of lung tuberculosis. Int J Comput Assist Radiol Surg. 2015;10:1–312.
Rahman T, Khandakar A, Kadir MA, Islam KR, Islam KF, Mazhar R, et al. Reliable tuberculosis detection using chest X-ray with deep learning, segmentation and visualization. IEEE Access. 2020;8:191586–601.
Duong LT, Le NH, Tran TB, Ngo VM, Nguyen PT. Detection of tuberculosis from chest X-ray images: boosting the performance with vision transformer and transfer learning. Expert Syst Appl. 2021;184: 115519.
Kiranyaz S, Ince T, Iosifidis A, Gabbouj M. Operational neural networks. Neural Comput Appl. 2020;32(11):6645–68.
kaggle. RSNA pneumonia detection challenge. 2020. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge . Accessed 9 Jun 2020.
Bustos A, Pertusa A, Salinas JM, De La Iglesia-Vaya M. Padchest: a large chest x-ray image dataset with multi-label annotated reports. Med Image Anal. 2020;66:101797.
Candemir S, Antani S. A review on lung boundary detection in chest X-rays. Int J Comput Assist Radiol Surg. 2019;14:563–76.
Gabrielian A, Engle E, Harris M, Wollenberg K, Glogowski A, Long A, Hurt DE, Rosenthal A. Comparative analysis of genomic variability for drug-resistant strains of Mycobacterium tuberculosis: the special case of Belarus. Infect Gene Evol. 2020;78:104137.
Long A, Glogowski A, Meppiel M, De Vito L, Engle E, Harris M, Ha G, Schneider D, Gabrielian A, Hurt DE, Rosenthal A. The technology behind TB DEPOT: a novel public analytics platform integrating tuberculosis clinical, genomic, and radiological data for visual and statistical exploration. J Am Med Inform Assoc. 2021;28(1):71–9.
Tahir AM, Chowdhury ME, Khandakar A, Rahman T, Qiblawey Y, Khurshid U, et al. COVID-19 infection localization and severity grading from chest X-ray images. Comput Biol Med. 2021;139: 105002.
Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017;2117–25.
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. Unet++: a nested U-Net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer; 2018. p. 3–11.
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention. 2015;234–41.
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017;4700–8.
Malik J, Kiranyaz S, Yamac M, Gabbouj M. Bm3d vs 2-layer Onn, in. IEEE International Conference on Image Processing (ICIP). 2021;2021:1994–8.
Malik J, Kiranyaz S, Gabbouj M. Self-organized operational neural networks for severe image restoration problems. Neural Net. 2021;135:201–11.
Malik J, Kiranyaz S, Gabbouj M. Operational vs convolutional neural networks for image denoising. ArXiv preprint; 2020. arXiv:2009.00612.
Malik J, Kiranyaz S, Gabbouj M. Image denoising by super neurons: why go deep? ArXiv preprint; 2021. arXiv:2111.14948.
Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KRK. Improvements to Platt’s SMO algorithm for SVM classifier design. Neural Comput. 2001;13(3):637–49.
Guo G, Wang H, Bell D, Bi Y, Greer K. KNN model-based approach in classification. OTM Confederated International Conferences On the Move to Meaningful Internet Systems. 2003;986–96.
Chen T, He T, Benesty M, Khotilovich V, Tang Y, Cho H, et al. Xgboost: extreme gradient boosting. R package version 04–2. 2015;1(4):1–4.
Pal M. Random forest classifier for remote sensing classification. Int J Remote Sens. 2005;26(1):217–22.
An T-K, Kim M-H. A new diverse AdaBoost classifier. Int Conf Artif Intell Comput Intell. 2010;2010:359–63.
Xanthopoulos P, Pardalos PM, Trafalis TB, Xanthopoulos P, Pardalos PM, Trafalis TB. Linear discriminant analysis. Robust Data Mining. 2013;27–33.
Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot. 2013;7:21.
Liao X, Xue Y, Carin L. Logistic regression with an auxiliary data source. In: Proceedings of the 22nd international conference on machine learning. 2005. p. 505–12.
Rahman T, Khandakar A, Islam KR, Soliman MM, Islam MT, Elsayed A, Qiblawey Y, Mahmud S, Rahman A, Musharavati F, Zalnezhad E. HipXNet: deep learning approaches to detect aseptic loosening of hip implants using X-ray images. IEEE Access. 2022;10:53359–73.
Rahman T, Khandakar A, Abir FF, Faisal MAA, Hossain MS, Podder KK, et al. QCovSML: a reliable COVID-19 detection system using CBC biomarkers by a stacking machine learning model. Comput Biol Med. 2022;143: 105284.
Rahman T, Ibtehaz N, Khandakar A, Hossain MSA, Mekki YMS, Ezeddin M, et al. QUCoughScope: an intelligent application to detect COVID-19 patients using cough and breath sounds. Diagnostics. 2022;12(4):920.
Smilkov D, Thorat N, Kim B, Viégas F, Wattenberg M. Smoothgrad: removing noise by adding noise. ArXiv preprint; 2017. arXiv:1706.03825.
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-cam: visual explanations from deep networks via gradient-based localization. Proc IEEE Int Conf Compu Vis. 2017;1:618–26.
Chattopadhay A, Sarkar A, Howlader P, Balasubramanian VN. Grad-cam++: generalized gradient-based visual explanations for deep convolutional networks. In 2018 IEEE Winter Conference on Applications of Computer Vision (WACV). 2018;2018:839–47.
Wang H, Wang Z, Du M, Yang F, Zhang Z, Ding S, et al. Score-CAM: score-weighted visual explanations for convolutional neural networks. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition workshops. 2020;24–5.
Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6(1):1–48.
TB QU dataset. https://www.kaggle.com/datasets/tawsifurrahman/tuberculosis-tb-chest-xray-dataset . Accessed 1 May 2022.
Meraj SS, Yaakob R, Azman A, Rum SNM, Shahrel A, Nazri A, et al. Detection of pulmonary tuberculosis manifestation in chest X-rays using different convolutional neural network (CNN) models. Int J Eng Adv Technol (IJEAT). 2019;9(1):2270–5.
Bhosale YH, Patnaik KS. IoT deployable lightweight deep learning application for COVID-19 detection with lung diseases using RaspberryPi. In 2022 International conference on IoT and blockchain technology (ICIBT). 2022;2022:1–6.
Yang F, Yu H, Kantipudi K, Karki M, Kassim YM, Rosenthal A, et al. Differentiating between drug-sensitive and drug-resistant tuberculosis with machine learning for clinical and radiological features. Quant Imaging Med Surg. 2022;12(1):675.
Ejiyi CJ, Qin Z, Nnani AO, Deng F, Ejiyi TU, Ejiyi MB, Agbesi VK, Bamisile O. ResfEANet: ResNet-fused external attention network for tuberculosis diagnosis using chest X-ray images. Comput Methods Programs Biomed Update. 2024;5:100133.
Liu CJ, Tsai CC, Kuo LC, et al. A deep learning model using chest X-ray for identifying TB and NTM-LD patients: a cross-sectional study. Insights Imaging. 2023;14:67.
Funding
Open Access funding provided by the Qatar National Library.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
This article is based on previously conducted studies and does not contain any studies with human participants or animals. The dataset used in this research is collected from several open access and restricted access datasets (upon request) where the standard protocols were used for creating the dataset.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rahman, T., Khandakar, A., Rahman, A. et al. TB-CXRNet: Tuberculosis and Drug-Resistant Tuberculosis Detection Technique Using Chest X-ray Images. Cogn Comput (2024). https://doi.org/10.1007/s12559-024-10259-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12559-024-10259-3