Abstract
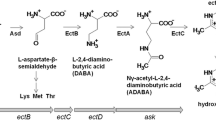
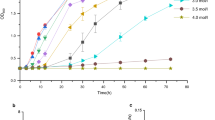
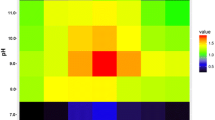
The moderately halophilic bacterium Halomonas sp. QHL1 was identified as a member of the genus Halomonas by 16S rRNA gene sequencing. HPLC analysis showed that strain QHL1 synthesizes ectoine in its cytoplasm. The genes involved in the ectoine biosynthesis pathway were identified on the chromosome in the order ectABC. Subsequently, the ectB gene from this strain was amplified by PCR, and the entire ectABC gene cluster (3,580 bp) was cloned using genome walking. Analysis showed that the ectA (579 bp), ectB (1269 bp), and ectC (390 bp) genes were organized in a single transcriptional unit and were predicted to encode three peptides of 21.2 kDa, 46.4 kDa, and 14.7 kDa, respectively. Two putative promoters, a δ70-dependent promoter and a δ38-controlled promoter, as well as several conserved motifs with unknown function were identified. Individual ectA, ectB, and ectC genes, and the entire ectABC gene cluster were inserted into the expression plasmid pET-28a(+) to generate the recombinant plasmids pET-28a(+)-ectA, pET-28a(+)-ectB, pET-28a(+)-ectC and pET-28a(+)-ectABC, respectively. Heterologous expression of these proteins in Escherichia coli BL21 (DE3) was confirmed by SDS-PAGE. The recombinant E. coli strain BL21 (pET-28a (+)-ectABC) displayed a higher salt tolerance than native E. coli cells but produced far less ectoine than the wild-type QHL1 strain.
Similar content being viewed by others
References
Bohnert, H.J., Nelson, D.E., and Jensen, R.G. 1995. Adaptations to environmental stresses. Plant Cell 7, 1099–1111.
Bordo, D., Monfort, R.L., Pijning, T., Kalk, K.H., Reizer, J., Saier, M.H., and Dijkstra, B.W. 1998. The three dimensional structure of the nitrogen regulatory protein IIANtr from Escherichia coli. J. Mol. Biol. 279, 245–255.
Brown, A.D. 1976. Microbial water stress. Bacteriol. Rev. 40, 803–846.
Calderón, M.I., Vargas, C., Rojo, F., Iglesias, G.F., Csonka, L.N., Ventosa, A., and Nieto, J.J. 2004. Complex regulation of the synthesis of the compatible solute ectoine in the halophilic bacterium Chromohalobacter salexigens DSM 3043T. Microbiology 150, 3051–3063.
Cánovas, D., Vargas, C., Calderón, M.I., Ventosa, A., and Nieto, J.J. 1998. Characterization of the genes for the biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halomonas elongata DSM 3043. System Appl. Microbiol. 21, 487–497.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Galinski, E.A., Pfeiffer, H.P., and Trüper, H.G. 1985. 1, 4, 5, 6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid: a novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur. J. Biochem. 149, 135–139.
Galinski, E.A. and Trüper, H.G. 1994. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol. Rev. 15, 95–108.
Graf, R., Anzali, S., Buenger, J., Pfluecker, F., and Driller, H. 2008. The multifunctional role of ectoine as a natural cell protectant. Clin. Dermatol. 26, 326–333.
Guzmán, H., Van-Thuoc, D., Martín, J., Hatti-Kaul, R., and Quillaguamán, J. 2009. A process for the production of ectoine and poly (3-hydroxybutyrate) by Halomonas boliviensis. Appl. Microbiol. Biotechnol. 84, 1069–1077.
Kuhlmann, A.U. and Bremer, E. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68, 772–783.
Kushner, D.J. and Kamekura, M. 1988. Physiology of halophilic bacteria, pp. 109–138. Halophilic bacteria, CRC Press Inc., Boca Raton, Florida, USA.
Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685.
Lee, S.J. and Gralla, J.D. 2004. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell 14, 153–162.
Lo, C.C., Bonner, A.C., Xie, G., D’Souza, M., and Jensen, R.A. 2009. Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways. Microbiol. Mol. Biol. Rev. 73, 594–651.
Louis, P. and Galinski, E.A. 1997. Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology 143, 1141–1149.
Maskow, T. and Babel, W. 2001. Calorimetrically obtained information about the efficiency of ectoine synthesis from glucose in Halomonas elongata. Biochim. Biophys. Acta. 1528, 60–70.
Miller, J.H. 1992. A laboratory manual and handbook for Escherichia coli and related bacteria, pp. 194–195. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, N.Y., USA.
Mustakhimov, I.I., Reshetnikov, A.S., Glukhov, A.S., Khmelenina, V.N., Kalyuzhnaya, M.G., and Trotsenko, Y.A. 2010. Identification and characterization of EctR1, a new transcriptional regulator of the ectoine biosynthesis genes in the halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. J. Bacteriol. 192, 410–417.
Nagata, S., Maekawa, Y., Ikeuchi, T., Wang, Y.B., and Ishida, A. 2002. Effect of compatible solutes on the respiratory activity and growth of Escherichia coli K-12 under NaCl stress. J. Biosci. Bioeng. 94, 384–389.
Oesterhelt, D. and Stoeckenius, W. 1974. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Meth. Enzymol. 31, 667–678.
Ono, H., Okuda, M., Tongpim, S., Imai, K., Shinmyo, A., Sakuda, S., Kaneko, Y., Murooka, Y., and Takano, M. 1998. Accumulation of compatible solutes, ectoine and hydroxyectoine, in a moderate halophile, Halomonas elongata KS3 isolated from dry salty land in Thailand. J. Ferm. Bioeng. 85, 362–368.
Onraedt, A.E., Walcarius, B.A., Soetaert, W.K., and Vandamme, E.J. 2005. Optimization of ectoine synthesis through fed-batch fermentation of Brevibacterium epidermis. Biotechnol. Prog. 21, 1206–1212.
Pastor, J.M., Salvador, M., Argandoña, M., Bernal, V., Reina-Bueno, M., Csonka, L.N., Iborraa, J.L., Vargasb, C., Nietob, J.J., and Cánovasa, M. 2010. Ectoines in cell stress protection: Uses and biotechnological production. Biotechnol. Adv. 28, 782–801.
Peters, P., Galinski, E.A., and Trüper, H.G. 1990. The biosynthesis of ectoine. FEMS Microbiol. Lett. 71, 157–162.
Reshetnikov, A.S., Khmelenina, V.N., Mustakhimov, I.I., Kalyuzhnaya, M., Lidstrom, M., and Trotsenko, Y.A. 2011. Diversity and phylogeny of the ectoine biosynthesis genes in aerobic, moderately halophilic methylotrophic bacteria. Extremophiles 15, 653–663.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Saum, S.H. and Müller, V. 2008. Growth phase-dependent switch in osmolyte strategy in a moderate halophile: ectoine is a minor osmolyte but major stationary phase solute in Halobacillus halophilus. Environ. Microbiol. 10, 716–726.
Schwibbert, K., Marin, S.A., Bagyan, I., Heidrich, G., Lentzen, G., Seitz, H., Rampp, M., Schuster, S.C., Klenk, H., Pfeiffer, F., Oesterhelt, D., and Kunte, H.J. 2011. A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581T. Environ. Microbiol. 13, 1973–1994.
Seip, B., Galinski, E.A., and Kurz, M. 2011. Natural and engineered hydroxyectoine production based on the Pseudomonas stutzeri ectABCD-ask gene cluster. Appl. Environ. Microbiol. 77, 1368–1374.
Severin, J., Wohlfarth, A., and Galinski, E.A. 1992. The predominant role of recently discovered tetrahydropyrimidines for the osmoadaptation of halophilic eubacteria. J. Gen. Microbiol. 138, 1629–1638.
Shikuma, N.J., Davis, K.R., Fong, J.N., and Yildiz, F.H. 2012. The transcriptional regulator, CosR, controls compatible solute biosynthesis and transport, motility and biofilm formation in Vibrio cholerae. Environ. Microbiol. 15, 1387–1399.
Springer, B., Kirschner, P., Rost-Meyer, G., Schröder, K.H., Kroppenstedt, R.M., and Böttger, E.C. 1993. Mycobacterium interjectum, a new species isolated from a patient with chronic lymphadenitis. J. Clin. Microbiol. 31, 3083–3089.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software Version 4.0. Mol. Biol. Evol. 24, 1596–1599.
Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Vargas, C., Argandona, M., Reina, B.M., Rodríguez, M.J., Fernández, A.C., and Nieto, J.J. 2008. Unravelling the adaptation responses to osmotic and temperature stress in Chromohalobacter salexigens, a bacterium with broad salinity tolerance. Saline Syst. 4, 14.
Ventosa, A., Nieto, J.J., and Oren, A. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62, 504–544.
Wang, Z., Ye, S., Li, J., Zheng, B., Bao, M., and Ning, G. 2011. Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol. 11, 109–121.
Wei, Y.H., Yuan, F.W., Chen, W.C., and Chen, S.Y. 2011. Production and characterization of ectoine by Marinococcus sp. ECT1 isolated from a high-salinity environment. J. Biol. Bioeng. 111, 336–342.
Woese, C.R., Gutell, R.R., Gupta, R., and Noller, H.F. 1983. Detailed analysis of the higher order structure of 16S-like ribosomal ribonucleic acids. Microbiol. Rev. 47, 621–669.
Wohlfarth, A., Severin, J., and Galinski, E.A. 1990. The spectrum of compatible solutes in heterotrophic halophilic eubacteria of the family Halomonadaceae. J. Gen. Microbiol. 136, 705–712.
Zhao, B., Lu, W., Yang, L., Zhang, B., Wang, L., and Yang, S.S. 2006. Cloning and characterization of the genes for biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halobacillus dabanensis D-8T. Curr. Microbiol. 53, 183–188.
Zhang, L.H., Lang, Y.J., and Nagata, S. 2009. Efficient production of ectoine using ectoine-excreting strain. Extremophiles 13, 717–724.
Zhu, D., Niu, L., Wang, C., and Nagata, S. 2007. Isolation and characterisation of moderately halophilic bacterium Halomonas ventosae DL7 synthesizing ectoine as compatible solute. Ann. Microbiol. 57, 401–406.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhu, D., Liu, J., Han, R. et al. Identification and characterization of ectoine biosynthesis genes and heterologous expression of the ectABC gene cluster from Halomonas sp. QHL1, a moderately halophilic bacterium isolated from Qinghai Lake. J Microbiol. 52, 139–147 (2014). https://doi.org/10.1007/s12275-014-3389-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-014-3389-5