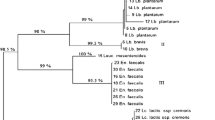
Abstract
A total of 152 lactic acid bacteria (LAB) were isolated from pickles produced in the Ankara-Çubuk region. These isolates were clustered into eight groups on the basis of their phenotypic characteristics including cell morphology, CO2 production from glucose, growth at 10 and 45 °C, growth in 6.5 % NaCl, and growth at pH 9.6. API 50 CH carbohydrate fermentation test, 16S ribosomal RNA (rRNA) sequence analysis, and sodium dodecyl sulfate-acrylamide gel electrophoresis (SDS-PAGE) whole-cell protein profile analysis were also performed for precise identification of the isolates at the species level. Molecular identification revealed that the most prevalent LAB species involved in pickle fermentation were Pediococcus ethanolidurans (46 isolates, 30.3 %), Lactobacillus brevis (37 isolates, 24.3 %), Lactobacillus plantarum (37 isolates, 24.3 %), and Lactobacillus buchneri (15 isolates, 9.9 %). Other LAB were found in minor frequencies such as Pediococcus parvulus (8 isolates, 5.3 %), Lactobacillus namurensis (6 isolates, 3.9 %), Lactobacillus diolivorans (1 isolate, 0.7 %), Lactobacillus parabrevis (1 isolate, 0.7 %), and Enterococcus casseliflavus (1 isolate, 0.7 %). When results of phenotypic and genotypic identification methods were compared, differences in the species distribution of LAB associated with pickles were defined between the API and the 16S rRNA sequencing. The API 50 CHL test coincided with the 16S rRNA results in 71 out of the 152 tested isolates, indicating that API gave unreliable identification results. A clear correlation could not be found between the results of whole-cell SDS profiles and 16S rRNA sequencing. Therefore, molecular characterization by 16S rRNA sequencing was considered to be the most reliable method for identifying isolates. The results presented in this work provide insight in to the LAB population associated with traditional Çubuk pickles and constitute a LAB strain resource for further studies involving the development of starter cultures.

Similar content being viewed by others
References
Axelsson L (2004) Lactic acid bacteria: classification and physiology. In: Salminen S, von Wright A, Ouwehand A (eds) Lactic acid bacteria: microbiological and functional aspects, 3rd edn. Marcel Dekker, New York
Balcázar JL, de Blas I, Ruiz-Zarzuela I, Vendrell D, Gironés O, Muzquiz JL (2007) Sequencing of variable regions of the 16S rRNA gene for identification of lactic acid bacteria isolated from the intestinal microbiota of healthy salmonids. Comp Immunol Microbiol Infect Dis 30:111–118
Boyd MA, Antonio MAD, Hillier SL (2005) Comparison of API 50 CH strips to whole-chromosomal DNA probes for identification of Lactobacillus species. J Clin Microbiol 43(10):5309–5311
Breidt F, Medina E, Wafa D, Pérez-Díaz I, Franco W, Huang H-Y, Johanningsmeier SD, Kim JH (2013) Characterization of cucumber fermentation spoilage bacteria by enrichment culture and 16S rDNA cloning. J Food Sci 78(3):M470–M476
Çetin B (2011) Production of probiotic mixed pickles (Turşu) and microbiological properties. Afr J Biotechnol 10(66):14926–14931
Chao S-H, Wu R-J, Watanabe K, Tsai Y-C (2009) Diversity of lactic acid bacteria in suan-tsai and fu-tsai, traditional fermented mustard products of Taiwan. Int J Food Microbiol 135:203–210
Chen Y, Wu H, Lo H, Lin W, Hsu W, Lin C, Lina P, Yanagida F (2012) Isolation and characterisation of lactic acid bacteria from jiang-gua (fermented cucumbers), a traditional fermented food in Taiwan. J Sci Food Agric 92:2069–2075
Çon AH, Karasu N (2009) Determination of antagonistic starter cultures for pickle and olive fermentation processes. Czech J Food Sci 27(3):185–193
De Angelis M, Corsetti A, Tosti N, Rossi J, Corbo MR, Gobbetti M (2001) Characterization of Non-starter lactic acid bacteria from Italian ewe cheeses based on phenotypic, genotypic, and cell wall protein analyses. Appl Environ Microbiol 67(5):2011–2020
Endo A, Mizuno H, Okada S (2008) Monitoring the bacterial community during fermentation of sunki, an unsalted, fermented vegetable traditional to the Kiso area of Japan. Lett Appl Microbiol 47:221–226
Franco W, Pérez-Díaz IM (2012) Role of selected oxidative yeasts and bacteria in cucumber secondary fermentation associated with spoilage of the fermented fruit. Food Microbiol 32:338–344
Franco W, Pérez-Díaz IM, Johanningsmeier SD, McFeeters RF (2012) Characteristics of spoilage-associated secondary cucumber fermentation. Appl Environ Microbiol 78(4):1273–1284
Hammes WP, Hertel C (2006) The genera Lactobacillus and Carnobacterium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The Prokaryotes A Handbook on the Biology of Bacteria, 3rd edn. Springer Science + Business Media, New York, pp 320–403
Hitchener BJ, Egan AF, Rogers PJ (1982) Characteristics of lactic acid bacteria isolated from vacuum-packaged beef. J Appl Bacteriol 52(1):31–37
Hutkins RW (2006) Microbiology and technology of fermented foods. Blackwell Publishing, USA
Iuchi A, Haruguchi S, Mongkolthanaruk W, Arima J, Nagase M, Khanh HQ, Ichiyanagi T, Yamaguchi T, Shimomura N, Aimi T (2012) Characterization of novel amylase from amylolytic lactic acid bacteria Pediococcus ethanolidurans isolated from Japanese pickles (nuka-zuke). Food Sci Technol Res 18(6):861–867
Johanningsmeier SD, McFeeters RF (2013) Metabolism of lactic acid in fermented cucumbers by Lactobacillus buchneri and related species, potential spoilage organisms in reduced salt fermentations. Food Microbiol 35:129–135
Kabak B, Dobson ADW (2011) An introduction to the traditional fermented foods and beverages of Turkey. Crit Rev Food Sci Nutr 51:248–260
Kato K, Toh H, Sakamoto N, Mori K, Tashiro K, Hibi N, Sonomoto K, Nakayamaa J (2014) Draft genome sequence of Lactobacillus namurensis Chizuka 01, isolated from nukadoko, a pickling bed of fermented rice bran. Genome Announc 2(1):e01263–13. doi:10.1128/genomeA. 01263-13
Kim S-Y, Adachi Y (2007) Biological and genetic classification of canine intestinal lactic acid bacteria and bifidobacteria. Microbiol Immunol 51(10):919–928
Kingston JJ, Radhika M, Roshini PT, Raksha MA, Murali HS, Batra HV (2010) Molecular characterization of lactic acid bacteria recovered from natural fermentation of beet root and carrot Kanji. Indian J Microbiol 50:292–298
Kizerwetter-Swida M, Binek M (2005) Selection of potentially probiotic Lactobacillus strains towards their inhibitory activity against poultry enteropathogenic bacteria. Pol J Microbiol 54(4):287–294
Krooneman J, Faber F, Alderkamp AC, Elferink SJHWO, Driehuis F, Cleenwerck I, Swings J, Gottschal JC, Vancanneyt M (2002) Lactobacillus diolivorans sp. nov., a 1,2-propanediol-degrading bacterium isolated from aerobically stable maize silage. Int J Syst Evol Microbiol 52:639–646
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, pp 115–147
Liu L, Zhang B, Tong H, Dong X (2006) Pediococcus ethanolidurans sp. nov., isolated from the walls of a distilled-spirit-fermenting cellar. Int J Syst Evol Microbiol 56:2405–2408
Markiewicz LH, Biedrzycka E, Wasilewska E, Bielecka M (2010) Rapid molecular identification and characteristics of Lactobacillus strains. Folia Microbiol 55(5):481–488
Moraes PM, Perin LM, Júnior AS, Nero LA (2013) Comparison of phenotypic and molecular tests to identify lactic acid bacteria. Braz J Microbiol 44(1):109–112
Pogačić T, Kelava N, Zamberlin Š, Dolenčić-Špehar I, Samaržija D (2010) Methods for culture-independent identification of lactic acid bacteria in dairy products. Food Technol Biotechnol 48(1):3–10
Pot B, Ludwig W, Kersters K, Schleifer K-H (1994) Taxonomy of lactic acid bacteria. In: De Vuyst L, Vandamme EJ (eds) Bacteriocins of lactic acid bacteria. Microbiology, genetics and applications. Springer Science + Business Media, New York, pp 13–90
Randazzo CL, Restuccia C, Romano AD, Caggia C (2004) Lactobacillus casei, dominant species in naturally fermented Sicilian green olives. Int J Food Microbiol 90:9–14
Reina LD, Breidt F Jr, Fleming HP, Kathariou S (2005) Isolation and selection of lactic acid bacteria as biocontrol agents for nonacidified, refrigerated pickles. J Food Sci 70(1):M7–M11
Sakamoto N, Tanaka S, Sonomoto K, Nakayama J (2011) 16S rRNA pyrosequencing-based investigation of the bacterial community in nukadoko, a pickling bed of fermented rice bran. Int J Food Microbiol 144:352–359
Sánchez I, Palop L, Ballesteros C (2000) Biochemical characterization of lactic acid bacteria isolated from spontaneous fermentation of ‘Almagro’ eggplants. Int J Food Microbiol 59:9–17
Sánchez I, Seseña S, Palop LL (2004) Polyphasic study of the genetic diversity of lactobacilli associated with ‘Almagro’ eggplants spontaneous fermentation, based on combined numerical analysis of randomly amplified polymorphic DNA and pulsed-field gel electrophoresis patterns. J Appl Microbiol 97:446–458
Scheirlinck I, Meulen RV, Schoor AV, Cleenwerck I, Huys G, Vandamme P, Vuyst LD, Vancanneyt M (2007) Lactobacillus namurensis sp. nov., isolated from a traditional Belgian sourdough. Int J Syst Evol Microbiol 57:223–227
Schillinger U, Holzapfel WH (1995) The genus Carnobacterium. In: Wood BJB, Holzapfel WH (eds) The genera of lactic acid bacteria, 1st edn. Chapman & Hall, UK, pp 307–324
Simpson WJ, Taguchi H (1995) The genus Pediococcus, with notes on the genera Tetratogenococcus and Aerococcus. In: Wood BJB, Holzapfel WH (eds) The genera of lactic acid bacteria, 1st edn. Chapman & Hall, UK, pp 125–172
Şimşek Ö, Çon AH, Tulumoğlu Ş (2006) Isolating lactic starter cultures with antimicrobial activity for sourdough processes. Food Control 17:263–270
Singh AK, Ramesh A (2008) Succession of dominant and antagonistic lactic acid bacteria in fermented cucumber: insights from a PCR-based approach. Food Microbiol 25:278–287
Tamang JP, Tamang B, Schillinger U, Franz CMAP, Gores M, Holzapfel WH (2005) Identification of predominant lactic acid bacteria isolated from traditionally fermented vegetable products of the Eastern Himalayas. Int J Food Microbiol 105:347–356
Tamminen M, Joutsjoki T, Sjöblom M, Joutsen M, Palva A, Ryhänen E-L, Joutsjoki V (2004) Screening of lactic acid bacteria from fermented vegetables by carbohydrate profiling and PCR–ELISA. Lett Appl Microbiol 39:439–444
Temmerman R, Huys G, Swings J (2004) Identification of lactic acid bacteria: culture-dependent and culture-independent methods. Trends Food Sci Technol 15:348–359
Vancanneyt M, Naser SM, Engelbeen K, Wachter MD, Meulen RV, Cleenwerck I, Hoste B, Vuyst LD, Swings J (2006) Reclassification of Lactobacillus brevis strains LMG 11494 and LMG 11984 as Lactobacillus parabrevis sp. nov. Int J Syst Evol Microbiol 56:1553–1557
Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J (1996) Polyphasic taxonomy, a consensus approach to bacterial systematic. Microbiol Rev 60(2):407–438
Xiong T, Guan Q, Song S, Hao M, Xie M (2012) Dynamic changes of lactic acid bacteria flora during Chinese sauerkraut fermentation. Food Control 26:178–181
Yu J, Gao W, Qing M, Sun Z, Wang W, Liu W, Pan L, Sun T, Wang H, Bai N, Zhang H (2012) Identification and characterization of lactic acid bacteria isolated from traditional pickles in Sichuan, China. J Gen Appl Microbiol 58:163–172
Zhang C, Brandt MJ, Schwab C, Gänzle MG (2010) Propionic acid production by cofermentation of Lactobacillus buchneri and Lactobacillus diolivorans in sourdough. Food Microbiol 27:390–395
Acknowledgments
This research was supported by the Scientific and Technological Research Council of Turkey (Project No. 108O491).
Author information
Authors and Affiliations
Corresponding author
Additional information
Simel Bağder Elmacı and Mehmet Tokatlı contributed equally to this work.
Rights and permissions
About this article
Cite this article
Bağder Elmacı, S., Tokatlı, M., Dursun, D. et al. Phenotypic and genotypic identification of lactic acid bacteria isolated from traditional pickles of the Çubuk region in Turkey. Folia Microbiol 60, 241–251 (2015). https://doi.org/10.1007/s12223-014-0363-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-014-0363-x