Abstract
The partitioning of reducing power into different electron-accepting pathways was evaluated during growth and stationary phases of H2-producing Rhodobacter sphaeroides cultures. For this, an electron balance method was developed using the chemical oxygen demand concept to quantitatively analyze the partitioning of nutrient electrons into H2, cell biomass, polyhydroxybutyrate (PHB), and soluble microbial products (SMP). Overall, these four electron sinks were accounted for greater than 85% of the electrons provided by the nutrients. Glucose, lactate, succinate, fumarate, and pyruvate were individually provided as the main carbon source, and in all cases, glutamate was provided as a nitrogen source in order to enhance H2 production. About 25–35% of the electrons ended up in H2 during growth, while up to 60% of the electrons partitioned into H2 in some stationary phase cultures. The other two major electron sinks in the growth phase were cell mass and PHB, while in stationary phase, SMP were accounted for >30% of the substrate electrons utilized. In general, the largest portion of SMP comprised low-molecular weight (<3 kDa) compounds mostly produced during stationary phase, although larger-size molecules were also detected in both phases. Overall, the fractions of electrons that partitioned into H2 (0.21 to 0.35) and PHB (0.06 to 0.21) were highly correlated with the standard free energy change of the substrate oxidation half-reaction equation, normalized per electron equivalent. In a PHB(−) mutant, electron redistribution increased H2 production, the extent of which depended on the carbon source provided.
Similar content being viewed by others
Introduction
Among the biologically based fuels that are being considered or developed as renewable replacements for fossil fuels are ethanol, various next generation liquid transportation additives, or gases like H2 or CH4 [1]. Strategies to achieve efficient biological routes for the production of these fuels require not only an understanding of the pathways by which cells partition the carbon skeletons and reducing power needed for fuel production, but also knowledge about the competing processes that detract from biofuel production. Using H2 as a model biofuel, we are interested in determining how cells partition available reducing power into different electron-accepting pathways. The goal of these studies is to identify competing pathways that can potentially be eliminated to optimize H2 production.
Biologically based H2-producing systems have the potential to reduce the amount of fossil fuels used for the fertilizer, chemical, petroleum, or steel industries, since H2 synthesis for their use accounts for approximately 10% of the national fossil fuel use [2]. Moreover, about 95% of the current global H2 is produced by individual industries to satisfy their own needs [3]. Such a decentralized production approach is an attractive model for the implementation of biological H2 production technologies.
The biological sources of H2 currently under consideration include both fermentative and photosynthetic organisms [4, 5]. Among the photosynthetic microbes already known to produce H2 are photoheterotrophic bacteria, which have the ability to simultaneously use energy derived from light and organic compounds to drive H2 production [6], and therefore, can be used to develop solar-driven biofuel production processes for the utilization of renewable organic substrates in a biofuel economy. In addition, the potential for recycling organic waste into H2 via photoheterotrophic metabolism [7, 8] or a combination of dark and light fermentations [9-11] has also been advocated, potentially leading to on-site reutilization of organic wastes by industries that currently use H2 produced from nonrenewable resources.
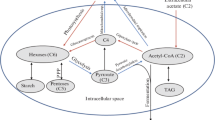
In this study, we quantitatively determined the fate of electrons in H2-producing batch cultures of the purple nonsulfur bacterium Rhodobacter sphaeroides during growth and stationary phases. R. sphaeroides is known to catalyze light- and nutrient-powered H2 production at significant rates [12-14]. Past studies of H2 production by this and other related bacteria have implicated nitrogenase activity as a source of this compound [6, 8, 15]. In addition, genetic and genomic techniques are available [16-18] and can be used to test predictive models for how individual pathways impact the flow of reducing power to H2. Based on prior knowledge on R. sphaeroides and other photoheterotrophic organisms [8, 19], a conceptual model of electron partitioning into pathways that contribute to and detract from H2 production is available (Fig. 1). Thus, our work sought to develop methods to assess the partitioning of reducing power into H2 production when wild type or mutant strains were provided light and different organic substrates.
Our approach is based on the theoretical or experimental assessment of the chemical oxygen demand (COD) of products from four electron-accepting reactions (Fig. 1). These products include new cells (biomass), polyhydroxybutyrate (PHB), soluble microbial products (SMP), and H2. Since COD is indicative of the reduction state of each product [20], this method allowed the quantitative determination of electron fate in a mass balance model (Fig. 1).
Methods
Bacterial Strains
All experiments were carried out using wild-type R. sphaeroides 2.4.1. or its derivative ∆0382, which is deficient in PHB synthesis. To construct ∆0382, a 5.4-kb fragment containing RSP0382 (annotated as coding for PHB synthase) plus 1.8-kb upstream and 1.7-kb downstream flanking DNA was amplified from the genome of strain 2.4.1 via polymerase chain reaction (PCR) using the upstream primer 5′-ACTCTAGACCTACGATTTCATGCCCGA-3′ (containing an XbaI restriction site, underlined) and the downstream primer 5′-TAGCATGCCGTCCGACTATTCCGAAG-3′ (containing an SphI restriction site, underlined). This fragment was digested with XbaI and SphI and cloned into XbaI- and SphI-digested pUC19 to generate plasmid pUC19/RSP0382. PCR amplification of this plasmid using the primers 5′-ATAGATCTTCGACCGACAAGACCTTCATTC-3′ and 5′-ATAGATCTTGCTCTTCGGTTGCCATATTGAC-3′ (both containing BglII restriction sites, underlined) was used to generate a 1,491-bp in-frame deletion within RSP0382. The resulting DNA fragment was digested with BglII, and then treated with T4 DNA ligase to produce plasmid pUC19/∆0382. This plasmid was digested with XbaI and SphI, and purified DNA containing the deletion in RSP0382 was cloned into XbaI- and SphI-digested pK18mobsacB (which contains kanamycin resistance and sucrose sensitivity (sacB) genes) to produce pK18mobsacB/∆0382. This plasmid was transformed into Escherichia coli S17-1 and mobilized via conjugation into R. sphaeroides 2.4.1. Kanamycin-resistant, sucrose-sensitive exconjugants, in which the plasmid was incorporated into the genome via homologous recombination, were isolated, and then grown in medium lacking kanamycin and sucrose. Sucrose-resistant, kanamycin-sensitive cells, in which the plasmid was excised via a second homologous recombination event, were isolated and screened by PCR for the deletion within RSP0382.
Growth Conditions and Sampling
Cultures were first grown aerobically in Sistrom's minimal medium [21] amended with 33.9 mM succinate as carbon source and ammonia as nitrogen source, and then under anaerobic photosynthetic conditions in a modified Sistrom's medium with 8.1 mM glutamate as the only nitrogen source and one of the following carbon sources: 39.5 mM lactate, 33.9 mM succinate, 39.5 mM fumarate, 47.4 mM pyruvate, or 19.8 mM glucose (concentrations were determined to meet a targeted constant COD of 3,800 mg/L for the carbon source). Cultures to be analyzed were inoculated by transferring 200 μL of photosynthetically grown cells into test tubes (∼21 mL total volume) with 20 mL of fresh medium containing the same carbon source.
Photosynthetic cells were grown at 30°C continuously illuminated by an incandescent light source that produced approximately 10 W/m2, as measured with a Yellow–Springs–Kettering model 6.5-A radiometer through a Corning 7–69 filter. Cell turbidity was monitored using a Klett–Summerson photoelectric colorimeter (Klett MFG Co., NY). Turbidity readings were converted to colony-forming units per milliliter using previously described conversion factors for photosynthetic cultures [22].
To measure the amount of gas produced, cultures were capped gastight with a rubber septum and the 1-mL headspace connected to an AER-200 respirometer (Challenge Technology, Springdale, AR) via a needle (≥20 G) and either 3/32-in. Tygon FTM tubing provided by the manufacturer or 2.06-mm Masterflex Viton extension tubing (Cole-Parmer, Vernon Hills, IL). The data acquisition and control software for the respirometer was set to anaerobic mode, during which, the number of bubbles passing through the unit was recorded in real time and converted to volume (milliliter) using factory-derived calibration constants.
Two or three cultures (biological replicates) were sampled at different phases of growth and gas production. For the analysis of gas composition, ∼3–7 mL of gas was removed from the culture headspace. Subsequently, 2 mL of culture was sampled and immediately frozen for total COD analysis, while the remaining culture was centrifuged (6,000 rpm, 12 min, 4°C). The supernatant was filtered through 0.2-μm syringe filters, and the filtered supernatant and cell pellets were immediately frozen and stored for analysis of SMP and PHB, respectively. When indicated, selected supernatants were passed through prewashed 10- or 3-kDa molecular weight cutoff filters (Millipore Microcon, Bedford, MA) for SMP size distribution analysis.
Analytical Methods
The gas composition of headspace samples was analyzed using a GC-2014 gas chromatograph (Shimadzu Scientific, Columbia, MD). A flame ionization detector (FID) (set at 200°C) coupled to a methanizer (380°C) and a thermal conductivity detector (100°C) were used for measuring CO2 and H2, respectively. The column oven temperature was 60°C. Argon was used as the carrier gas. For quantification, a standard of 80% H2 and 20% CO2 (Airgas, Carol Stream, IL) and a Scotty 48 standard (Scotty Specialty Gases, Plumsteadville, PA) were used. The FID–methanizer system was capable of detecting volatile organic compounds such as ethylene, formic acid, and ethanol, although no significant amounts of these compounds were observed from the culture headspace samples tested (data not shown). Peaks for H2, CO2, O2, and N2 gases were typically detected, with the latter two being derived from small amounts of air introduced during the processing of samples from the cultures. A correction was therefore applied to calculate the H2 partial pressure in the cultures by removing the contributions of O2 and N2. The amount of H2 produced was calculated as the average H2 partial pressure during a batch times the total gas volume quantified by the respirometer.
Organic acids in culture supernatants were quantified by gas chromatography–mass spectrometry (GC–MS; GC-2010 gas chromatograph coupled to a QP-2010S mass spectrometer detector; Shimadzu Scientific) after derivatization with methyl chloroformate [23] and using benzoic acid methyl ester (BAME) as an internal standard. The carrier gas was helium, and the GC column was a SHRXI-5MS (Shimadzu Scientific). The temperatures for column oven, injection, ion source, and interface were set at 60°C, 180°C, 220°C, and 275°C, respectively. Standards of known concentrations were used to generate calibration curves for the quantification of identified peaks.
Sugar analysis was performed by high-performance liquid chromatography (model 10A-vP equipped with an RID-10A refractive index detector, Shimadzu Scientific) using operational settings established by the National Renewable Energy Laboratory [24]. Separation was achieved with an HPX-87P column (Biorad, Hercules, CA) set at 80°C, and detection was by refractive index at the same temperature. The mobile phase was 0.005 M H2SO4. Sugar standards were used for the generation of standard curves.
The PHB content of cells was determined following established procedures [25, 26] with slight modifications. Briefly, freeze-dried cell pellets were weighed, digested overnight in acidified methanol and chloroform at 105°C, mixed with deionized water to induce phase separation, and analyzed by GC–MS. Column oven and injection temperatures were set at 50°C and 210°C, respectively. PHB quantification was based on a PHB standard (Sigma-Aldrich, Steinheim, Germany) and with BAME as an internal standard. The fraction of PHB in biomass was calculated based on the total sample dry weight prior to digestion.
Electron Balance Calculations
COD was used to quantify electron availability and partitioning in gaseous, soluble, and particulate electron sinks. Theoretical COD values of substrates, H2, and PHB were calculated using Eq. 1 [20]:
The stoichiometry in this reaction defines the theoretical oxygen demand for the full oxidation of an organic compound to CO2. For example, the moles of O2 required for the complete oxidation of 1 mol of lactic acid (C3H6O3) can be calculated from Eq. 1 to be equal to 3 mol of O2. Since the molecular weight of O2 is 32 mg/mmol, the theoretical COD of lactic acid is 96 mg COD per millimole. Since the lactic acid experiments had an initial concentration of 39.5 mM, this corresponds to 3,800 mg COD per liter. Glutamic acid (C5H9O4N), the nitrogen source in all experiments, also contributes to the initial COD in the cultures. From Eq. 1 follows that 4.5 mol O2 is required for the complete oxidation of 1 mol of glutamic acid. Thus, the theoretical COD of glutamic acid is 144 mg COD per millimole, and since 8.1 mM of glutamic acid was used in all experiments, this corresponds to approximately 1,200 mg COD per liter. Consequently, the targeted initial COD in the medium, for all experiments, was about 5,000 mg/L. For PHB, a theoretical COD of 144 mg COD per millimole of monomer (C4H6O2) was calculated. For H2, the theoretical COD is 16 mg COD per millimole.
For mixed or unknown organic material, the COD cannot be calculated from Eq. 1, but can be obtained experimentally. Thus, the COD of the biomass and culture supernatants were determined colorimetrically using HACH High Range (0–1,500 mg/L) COD kits (HACH Company, Loveland, CO). Standard curves were prepared with potassium phthalate, according to standard methods [27].
Results
To understand electron partitioning during growth and stationary phases in R. sphaeroides, we used sets of batch cultures incubated under identical conditions, with two or three cultures (i.e., biological replicates) sampled at different stages of incubation. Real-time monitoring of gas production and high-frequency sampling of cell growth were implemented in each batch culture. Individual growth curves were fit to a single Gompertz sigmoid function (cell density = a (exp (b (exp (ct)))) [28], where a, b, and c are functional constants, and t is time) by adjusting the time variable and finding a unique set of constants that defined a characteristic shape of the growth curve for the entire set of cultures. The best-fitting sigmoid curves were in good agreement with experimental data (R 2 ≥ 0.99), which meant that individual growth profiles had similar shapes, and thus, the fitting could be used to synchronize these individual profiles within the same time frame, irrespective of the phase differences between cultures (i.e., because of variations in the lag phase). This procedure allowed the definition of five specific points in the growth curve in which electron partitioning was analyzed. These sampling points are illustrated in Fig. 2a, which shows growth and gas production results for a lactate-fed set of batch cultures. The first sampling point was at the onset of gas production and coincided with midexponential growth (designated as me). The second was shortly after the transition from exponential to arithmetic growth and was defined as a postexponential (pe) sampling point. The third sampling point was at the beginning the stationary (st) phase and was characterized by having the maximum gas production rate (Fig. 2b). Samplings in the middle of the stationary (mst) phase and at the end of batch (eob), when gas production stopped, completed the set of sampling points.
Time series of batch cultures grown with lactate. a Gas production and absorbance values showing cell growth. Curve shows best-fitting sigmoid function (cell density = 594exp (−13.6exp (−0.06t))) which is used for aligning time frames of different batch cultures in the set. Sampling points are indicated by arrows. Regions of interest are indicated with horizontal brackets. Inset shows values in logarithmic scale. See also the list of abbreviations. b Gas production rates shown in milliliter per hour and in milliliter per hour per cell. Normalization by number of cells was based on the best-fitting figure in a, with Klett unit converted to cells according to 1 Klett = 107 cells [22]. c Time series of lactate, glutamate, and total soluble compounds all expressed in milligram per liter oxygen demand. The difference of soluble COD (sCOD) from the sum of lactate and glutamate gives the COD content of SMP. Error bars standard deviations
At each of the sampled points, the concentrations of extracellular carbon and nitrogen sources were measured and converted to COD to quantify the reducing power remaining and consumed at different stages of growth. The COD profiles of the lactate-grown culture (Fig. 2c) indicated that about two thirds of lactate electrons had been utilized by the end of the growth phase (st point), and that the end of growth corresponded to the point where glutamate, the nitrogen source, was nondetectable. The remaining lactate was consumed by the cells during stationary phase. The difference between the measured soluble COD in the media and COD from the remaining carbon and nitrogen sources corresponded to SMP accumulation (see below for SMP analysis). In the case of lactate, SMP accumulation occurred mostly during stationary phase (Fig. 2c).
In addition to soluble COD, H2 evolved from the culture and biomass accumulation was quantified at each sampling point. For the calculation of COD from H2, the gas composition was measured at regular intervals, and an average for the entire experiment was derived (e.g., H2 represented 87 ± 3% of the evolved gas in lactate-fed cultures). The PHB component of the biomass was quantified separately, to complete COD calculations for all four major electron sinks depicted in Fig. 1. The resulting electron partitioning at each sampling point is illustrated in Fig. 3 for the lactate-fed culture. The COD sum of the end products is compared to the total substrate COD utilized (i.e., lactate and glutamate consumed) and the difference indicated as “CODother” in Fig. 3, which represents potentially unaccounted electron sinks or the accumulation of experimental error from individual products. As shown in Fig. 3, the cumulative amount of electrons directed toward H2 production continuously increased, with 55% of the total produced during the growth phase and the rest in the stationary phase. Cell synthesis and PHB were important electron sinks only during the growth phase, while the importance of SMP in the electron balance significantly increased in the stationary phase. Furthermore, CODother was less than 15% of the utilized COD for all time points, indicating that we can quantitatively account for most of the electrons derived from media components by measuring the products included in the study.
Time series of COD distribution in lactate-fed batch cultures. Sampling points are indicated by abbreviations in Fig. 2 a. Values greater than 200 mg/L are indicated on the bars. Error bars standard deviations. See also the list of abbreviations
Electron Partitioning with Different Carbon Sources
Electron partitioning in cultures fed with different carbon sources are compared in Fig. 4, where COD values have been normalized dividing by the COD consumed at the end of each experiment. Since, in all cases, the total COD consumption was virtually equal to the sum of initial COD in glutamate and in the carbon source, the electron partitioning in Fig. 4 represents essentially all available electrons from the media components. The data shows that, in all cases, more than 85% of substrate electrons was partitioned into the known electron sinks (i.e., fother < 0.15 in all cases). Succinate- and lactate-fed cultures had the highest partitioning into H2, while glucose-fed cultures produced significantly lower H2 than the organic acids tested. This trend was inversely correlated with PHB production, which was highest in glucose- and lowest for succinate- or lactate-fed cultures. An unexpected result was the significant fraction of electrons partitioning into SMP by the end of each experiment.
In addition to the end point analysis shown in Fig. 4, a comparison of electron fate during growth and stationary phases of the batch cultures is presented in Fig. 5. The phases are defined in Fig. 2a, as the postexponential growth phase (PEGP) and the stationary phase (STP). Notably, PEGP was characterized by a somewhat steady rate of H2 production at the single cell level, as the rate of gas evolution per cell and the percentage of H2 in the gas train did not vary significantly during this period (Fig. 2b). The electron partitioning in each phase was calculated by subtracting the COD values at the beginning point of each region from those at the end, and standard deviations were recalculated using standard error propagation techniques. During PEGP (Fig. 5a), biomass production was the main electron sink, followed by H2 and PHB production. During STP (Fig. 5b), biomass formation was generally negligible, and the main electron sinks were H2 and SMP. One exception to this generalization is the relatively large fraction of electrons accumulated in PHB by stationary phase cells in the glucose-fed culture (Fig. 5b), while in cultures fed with organic acids, net formation of PHB occurred only during growth (Fig. 5a, b). The negative values in Fig. 5 indicate product consumption rather than formation, and therefore, a general trend in the stationary phase was the utilization of small amounts of biomass and PHB as electron donors, except for the glucose culture, as explained above. Also, the net loss of SMP in PEGP by the pyruvate-fed culture (Fig. 5a) indicates a large production of SMP before the initiation of PEGP in this particular culture.
COD distribution at the regions of interest shown in Fig. 2a. a Postexponential growth phase. b Stationary phase. Values greater than 10 mg COD per liter are indicated on the bars. Error bars standard deviations calculated by error propagation. See also the list of abbreviations
Table 1 provides a quantitative comparison of gas accumulation, composition, and production rates for all experiments. The biogas composition was similar in all cultures (average H2 content of 87 ± 3%), but the amount of gas accumulated and the gas production rates per cell depended highly on the substrate. When the fraction of electrons going to H2 at each phase is normalized with respect to the amount of electron donors consumed (fH2 in Table 1), the organic acid-fed cultures show a higher utilization of H2 as electron sink in stationary phase than in the growth phase, but the glucose-fed culture shows by far the smallest partitioning of electrons into H2 in stationary phase, which correlates with the observed PHB accumulation during stationary phase in this culture. Thus, from the perspectives of maximizing H2 production, PHB formation seems to be an important competing electron sink during growth phase in all the cultures, but in the glucose-fed culture, PHB is also a competing electron sink during stationary phase.
Electron Partitioning in PHB(−) Mutants
Given the importance of PHB in the electron partitioning during growth and stationary phases, the effect of eliminating PHB as an electron sink was investigated with lactate- or glucose-fed cultures using electron balances for the overall batch (Fig. 4) and separately analyzing electron partitioning in PEGP and STP (Fig. 5). Overall electron balances in the PHB(−) mutant (Fig. 4) showed that electrons were redistributed among several other products, including H2, SMP, and cells. In the lactate-fed PHB(−) mutant culture, the increase in the electron share of H2 (from 34% to 37%) was small but significant at p = 0.05 (based on a Student's t test) and reproducible in independent experiments with lactate (data not shown), whereas the gain in electrons partitioning to H2 in the PHB(−) mutant was comparatively larger for glucose-fed cells (21% to 26%) with a p = 0.01, consistent with the electron partitioning to PHB being larger with glucose-fed wild type cells.
Analysis of SMP
Since the SMP pool was significant in all the cultures, we further analyzed SMP in two ways. Firstly, we quantified the overall size profile of the COD represented in SMP by filtering the soluble portion of cultures through 3- and 10-kDa filters. Secondly, we identified and quantified the low molecular weight, extracellular organic acids during PEGP and STP (Fig. 6).
Analysis of SMP formed and/or consumed during the batch tests. Left panel Size distribution analysis of SMP in postexponential growth phase (a) and stationary phase (b). Right panel Quantification of identified organic acids in postexponential growth phase (c) and stationary phase (d). Error bars standard deviations calculated by error propagation. See also the list of abbreviations
In general, there was significant variability in the size distribution of SMP between batch experiments with different carbon sources (Fig. 6). The data indicate that the SMP produced during STP was mostly <3-kDa molecules (Fig. 6b), while the size distribution was more dispersed during the PEGP (Fig. 6a). Macromolecules larger than 10 kDa were produced by most cultures during both growth phases and always showed net average production, suggesting little or no degradation of this SMP class. However, smaller size SMP was both produced and consumed.
SMP identified in the <3-kDa fraction included tricarboxylic acid (TCA) cycle intermediates, pyruvate and lactate (Fig. 6c, d). These generally did not make a large fraction of the <3-kDa portion, except in the glucose-fed culture of the PHB(−), in which, pyruvate was a major product during PEGP and was completely consumed during STP. For the other cultures, the <3-kDa SMP mostly consisted of unknown molecules, although moderate amounts of pyruvate, malate, and lactate accumulated in PEGP and were consumed in STP. The large amount of SMP consumed by the growing pyruvate culture and characterized as <3 kDa (Fig. 6a) remained largely unidentified, as there was no consumption of identified molecules during PEGP. Finally, succinate emerged as a common SMP produced during STP, although in moderate amounts (Fig. 6d).
Discussion
While photosynthetic microbes are among those being considered for biofuel production, little is known about the fate of electrons derived from light- or nutrient-driven metabolic processes. Therefore, it is desirable to develop approaches to quantify the amount of reducing power that is directed into fuels and major microbial end products and to use this information to increase the production of biofuels. In this study, we applied COD-based approaches to quantify electron fate during H2 production by the photosynthetic microbe R. sphaeroides. However, the assays and approaches described in this paper are applicable to other types of biofuel production by this and other microbes provided that electron donors and acceptors are either directly converted to COD units, or indirectly quantified in COD units by measuring products of their reactions.
During anoxic growth of photoheterotrophic organisms, reduced compounds present in the growth medium are the major source of electrons, as light energy capture is a cyclic process with no net electron gain [29]. Thus, by measuring the gaseous, liquid, and solid products of photoheterotrophic activity (Fig. 1), we were able to calculate an overall mass balance of electrons after converting the measurements to the common unit of COD. The unaccounted proportion of electrons (i.e., fother in Fig. 1) was generally 0–15%, indicating that the electron sinks considered in our conceptual model were the major ones.
Comparison of H2 Production by Growing and Resting Cells
H2 production by cultures of photoheterotrophic bacteria has been explored using resting or immobilized cell cultures [14, 30-32] to eliminate biomass formation as a competing reaction. However, to develop continuous flow H2-producing bioreactors [33], a growing culture is needed so biomass is a significant competing electron sink. By comparing electron partitioning analysis during growth and stationary phases (i.e., PEGP and STP regions defined in Fig. 2), our analysis provides a direct comparison of biofuel production efficiency during each stage (Fig. 5). It is evident from our results that the efficiency of fuel production depends not only on the growth conditions, but also on the ultimate aim of the process to be engineered. For instance, although the highest rate of H2 production is achieved at the end of exponential growth (Fig. 2b), electron partitioning into this fuel is still relatively low at this stage (Table 1) since biomass production is a major competitor. In contrast, the fraction of electrons routed to H2 is higher in stationary phase, while production rates follow a decreasing trend as one or more nutrients becomes limiting. Thus, if the goal is to maximize H2 production rates, flow-through bioreactors with a capacity for high cell concentrations might be desirable, even though light penetration might be a concern with photosynthetic microbes [33, 34]. However, if the goal is to maximize the conversion of one or more renewable substrates to H2, immobilized nongrowing cultures are predicted to be optimal [30, 32]. Finally, if the aim is to couple H2 production with either waste treatment [10] or removal of organic substrates from a liquid stream [35], then efficient substrate consumption in a plug flow reactor might provide the best results [33]. In that case, the electron balance of the entire experiment, including growth and stationary phases, may be a better guide for process design, although the discovery that a significant fraction of electrons can be partitioned to SMP suggests a limit in the removal efficiency of organic substrates.
Correlations of H2 and PHB Production with Free Energy of Organic Substrates
Analysis of our data allows two noteworthy correlations regarding electron partitioning in various cultures, which could potentially lead to a new approach for predicting H2 and PHB formation in photoheterotrophic cultures as a function of the organic substrate. Firstly, the fraction of electrons partitioned into H2 (Fig. 4) inversely correlate with the free energy changes of the electron donor half-reactions, on a per electron equivalent basis (r = −0.97, p < 0.01; Fig. 7). These free energy values were calculated based on free energies of formation of individual species [36] for a stoichiometrically balanced half-reaction normalized per unit of electrons transferred [20]. The correlation suggests that electron partitioning into H2 could be predicted by the free energy change in the degradation of the substrate to a common oxidized intermediate, rather than the initial oxidation state of the substrate. Indeed, while lactate and glucose have the same oxidation state, they were on the opposite ends of the H2 production spectrum. Secondly, the fraction of electrons going to PHB is also related to free energy changes of the substrate half-reactions (Fig. 7), in this case with a positive correlation (r = 0.99, p < 0.01). Interestingly, the sum of fH2 and fPHB was nearly constant (0.42 ± 0.02) over the five different carbon sources, as can be inferred from the slopes of H2 and PHB lines, whose absolute values are almost identical (Fig. 7). Thus, the inverse relationship in Fig. 7 clearly shows the competing nature of PHB and H2 formation for substrate electrons, although the mechanisms behind this competition and the free energy relationships are not clear. In contrast, there was no obvious correlation between SMP production and substrate free energy (r = 0.25, p > 0.65), possibly reflecting the diverse chemical nature of the SMP pool observed with different organic substrates (Fig. 6).
Correlation of free energy changes of electron donor half-reactions with electron partitioning into H2 and PHB. Free energy values are calculated for pH = 7 and normalized to one-electron reactions [20]. Arrows show the increase in electron partitioning into H2 upon PHB mutation. The positions of carbon sources on the free energy line are indicated on the upper axis. Error bars standard deviations. See also the list of abbreviations
Effects of Genetic Changes on the Distribution of Electrons
The strategy employed in this study is also suited for testing the predictions on the effects of removing individual electron sinks on production of H2 or other candidate fuel molecules. While PHB mutants of photoheterotrophic organisms have been used for increased H2 production before [37-40], little is known about how cells balance electron flux in these or other mutant strains. Our data indicate that loss of PHB synthesis removed a significant electron sink. However, the magnitude of the effect of blocking PHB synthesis on biofuel production depended on the substrate being utilized. For lactate, the net gain in H2 production by the PHB mutation was 7%, compared to H2 production in wild-type cultures (Table 1). Overall, the electron flux that was deviated from PHB was captured in H2 production (33% of electron flux), cell synthesis (44%), and SMP (22%). For glucose, there was a 30% higher H2 production in the PHB(−) mutant (Table 1). However, only 24% of the electron flux originally going to PHB in the wild-type cells contributed to increased H2 production in the PHB(−) mutant, as SMP was the most significant electron sink, receiving 52% of the deviated electron flux. The remaining flux corresponded to cell synthesis (9.5%) and an increase in the unaccounted CODother fraction.
Electron partitioning of the PHB(−) mutant was also analyzed during PEGP and STP (Fig. 5). In the lactate-fed PHB(−) mutant culture, the absolute values of H2 COD, as compared to those of the wild type, did not reflect the elimination of PHB production during growth and lack of electrons from PHB degradation during stationary phase (Fig. 5). This can be explained by considering the different amounts of total COD utilized during PEGP and STP in the cultures. When the total COD was used to normalize data, the fraction of H2 electrons in PHB(−) mutant was more than that in wild type during PEGP and less during STP (Table 1), as expected.
In the growth phase of the glucose-fed mutant cultures, there were approximately twofold increases in gas production rate and in H2 accumulation compared to glucose-fed wild-type cultures (Fig. 5a, Table 1). Accordingly, the fraction of glucose utilized during PEGP by the PHB(−) mutant increased significantly. During the stationary phase, the PHB(−) mutant yielded less H2 (Fig. 5b), despite the fact that PHB formation was a significant competing electron sink for the wild-type culture in this phase (Fig. 5b). This decrease is consistent with substrate utilization during growth, as less glucose was available at the initiation of the stationary phase in the PHB(−) mutant culture.
The increased H2 production in the PHB(−) mutant cultures was comparable to H2 production by wild-type cultures fed with pyruvate or fumarate (Table 1). This is important since biofuel production processes might use mixed organic acids for photoheterotrophic H2 production, following a dark fermentation step [9-11]. In this case, the benefit of utilizing a PHB(−) mutant would be small, since H2 production by wild-type cells is already high when fed organic acid substrates. On the contrary, the benefit of eliminating PHB accumulation would be more important during direct photoheterotrophic conversion of sugars to H2, because of the lower H2 production from sugars by the wild-type cells.
SMP as a Major Electron Sink
A surprising finding in this study was the observation that SMP can be a major electron sink, thus taking a large amount of reducing power away from biofuel production, especially in resting cells. SMP accumulation was shown to be considerably larger than PHB formation, and therefore, a logical target for further attempts to optimize H2 production would be the control of genetic and metabolic systems that lead to SMP production. Under the conditions described in this study, only a small fraction of the SMP corresponded to common metabolites such as lactate and TCA cycle intermediates (Fig. 6), and most remained unidentified low or high (>3 kDa) molecular weight material. In general, there is a fundamental lack of knowledge regarding SMP formation during photoheterotrophic growth, and in addition to the products identified in this study, formate is the only other organic acid that has been described as a byproduct of R. sphaeroides photoheterotrophic metabolism [33, 41], although this compound was not detected in any of the cultures in this study.
It is possible that the large fraction of high-molecular weight unidentified SMP could be extracellular polymeric substances (EPS), which are known to play a major role in cell aggregation [42, 43]. Although cell aggregation did not seem to occur to a significant extent in growing and early stationary phase cultures, there was noticeable cell sedimentation in late stationary phase, consistent with the possibility of EPS formation. Thus, the >10-kDa SMP observed in the cultures could possibly correspond to EPS precursors, a hypothesis that requires further investigation.
Conclusions
In conclusion, a COD-based method was developed and used for the quantitative assessment of electron partitioning in H2-producing R. sphaeroides cultures. This approach is useful for evaluating biofuel production by wild-type and mutant cultures in both growing and resting cells. The results with selected organic substrates revealed correlations of H2 production and PHB accumulation with the free energy for oxidation of the substrate, normalized on an electron equivalent basis. We also showed that the method is applicable to the analysis of the redistribution of reducing power, when a known electron sink, PHB synthesis, is removed by genetic modification. SMP was a significant electron sink competing with biofuel production, so future studies should target identification and removal of the pathways that will divert electrons from H2 or other desired biofuel products to SMP. Finally, the data generated on the distribution of reducing power for growing and resting cells can be useful to inform genome-scale metabolic network models of R. sphaeroides that will provide predictions on the flow of reducing power needed to further improve biofuel production.
Abbreviations
- BAME:
-
Benzoic acid methyl ester
- COD:
-
Chemical oxygen demand
- eob:
-
End of batch
- EPS:
-
Extracellular polymeric substances
- FID:
-
Flame ionization detector
- Fum:
-
Fumarate
- Glc:
-
Glucose
- Lac:
-
Lactate
- me:
-
Midexponential point
- mst:
-
Midstationary point
- pe:
-
Beginning of postexponential phase
- PEGP:
-
Postexponential growth phase
- PHB:
-
Polyhydroxybutyrate
- Pyr:
-
Pyruvate
- SMP:
-
Soluble microbial products
- st:
-
Beginning of stationary phase
- STP:
-
Stationary phase
- Suc:
-
Succinate
- WT:
-
Wild type
References
Demirbas A (2008) Biofuels sources, biofuel policy, biofuel economy and global biofuel projections. Energy Convers Manage 49:2106–2116
US Department of Energy (2008) A decade of discovery. http://www.energy.gov/discovery/
Mueller-Langer F, Tzimas E, Kaltschmitt M, Peteves S (2007) Techno-economic assessment of hydrogen production processes for the hydrogen economy for the short and medium term. Int J Hydrogen Energy 32:3797–3810
Das D, Veziroglu TN (2001) Hydrogen production by biological processes: a survey of literature. Int J Hydrogen Energy 26:13–28
Hallenbeck PC, Ghosh D (2009) Advances in fermentative biohydrogen production: the way forward? Trends in Biotechnology 27:287–297
Gest H, Kamen M (1949) Photoproduction of molecular hydrogen by Rhodospirillum rubrum. Science 109:558–559
Kapdan IK, Kargi F (2006) Bio-hydrogen production from waste materials. Enzyme Microb Technol 38:569–582
Rey FE, Heiniger EK, Harwood CS (2007) Redirection of metabolism for biological hydrogen production. Appl Environ Microbiol 73:1665–1671
Chen CY, Yang MH, Yeh KL, Liu CH, Chang JS (2008) Biohydrogen production using sequential two-stage dark and photo fermentation processes. Int J Hydrogen Energy 33:4755–4762
Eroglu E, Eroglu I, Gunduz U, Turker L, Yucel M (2006) Biological hydrogen production from olive mill wastewater with two-stage processes. Int J Hydrogen Energy 31:1527–1535
Nath K, Kumar A, Das D (2005) Hydrogen production by Rhodobacter sphaeroides strain OU001 using spent media of Enterobacter cloacae strain DM11. Appl Microbiol Biotechnol 68:533–541
Fascetti E, Todini O (1995) Rhodobacter sphaeroides RV cultivation and hydrogen production in a one- and two-stage chemostat. Appl Microbiol Biotechnol 44:300–305
Koku H, Eroglu I, Gunduz U, Yucel M, Turker L (2003) Kinetics of biological hydrogen production by the photosynthetic bacterium Rhodobacter sphaeroides OU 001. Int J Hydrogen Energy 28:381–388
Zhu HG, Suzuki T, Tsygankov AA, Asada Y, Miyake J (1999) Hydrogen production from tofu wastewater by Rhodobacter sphaeroides immobilized in agar gels. Int J Hydrogen Energy 24:305–310
Cho YK, Donohue TJ, Tejedor I, Anderson MA, McMahon KD, Noguera DR (2008) Development of a solar-powered microbial fuel cell. J Appl Microbiol 104:640–650
Callister SJ, Dominguez MA, Nicora CD, Zeng XH, Tavano CL, Kaplan S et al (2006) Application of the accurate mass and time tag approach to the proteome analysis of sub-cellular fractions obtained from Rhodobacter sphaeroides 2.4.1. aerobic and photosynthetic cell cultures. Journal of Proteome Research 5:1940–1947
Donohue TJ, Kaplan S (1991) Genetic techniques in Rhodospirillaceae. Methods Enzymol 204:459–485
Mackenzie C, Choudhary M, Larimer FW, Predki PF, Stilwagen S, Armitage JP et al (2001) The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth Res 70:19–41
Koku H, Eroglu I, Gunduz U, Yucel M, Turker L (2002) Aspects of the metabolism of hydrogen production by Rhodobacter sphaeroides. Int J Hydrogen Energy 27:1315–1329
Rittmann BE, McCarty PL (2001) Environmental biotechnology: principles and applications. McGraw-Hill, Boston
Sistrom W (1960) A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J Gen Microbiol 22:778–785
Tai SP, Kaplan S (1985) Intracellular localization of phospholipid transfer activity in Rhodopseudomonas sphaeroides and a possible role in membrane biogenesis. J Bacteriol 164:181–186
Villas-Boas SG, Delicado DG, Akesson M, Nielsen J (2003) Simultaneous analysis of amino and nonamino organic acids as methyl chloroformate derivatives using gas chromatography–mass spectrometry. Anal Biochem 322:134–138
Siluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D (2006) Determination of sugars, byproducts, and degradation products in liquid fraction process samples, vol. Technical Report: NREL/TP-520-42623. National Renewable Energy Laboratory
Comeau Y, Hall KJ, Oldham WK (1988) Determination of poly-beta-hydroxybutyrate and poly-beta-hydroxyvalerate in activated-sludge by gas–liquid chromatography. Appl Environ Microbiol 54:2325–2327
Drnevich MA, Noguera DR (2004) Production of polyhydroxyalkanoate during treatment of low-phosphorus-content wastewater. Water Environ Res 76:380–383
American Public Health Association (1989) 5220D Closed Reflux, calorimetric method. In: Clesceri LS, Greenberg AE, Trussell RR (eds) Standard methods for the examination of water and wastewater, 17th edn. American Public Health Association, Washington, DC, pp 5-15–5-16
Gompertz B (1825) On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. Phil Trans Roy Soc London 115:513–585
Johnson ET, Schmidt-Dannert C (2008) Light-energy conversion in engineered microorganisms. Trends in Biotechnology 26:682–689
Gosse JL, Engel BJ, Rey FE, Harwood CS, Scriven LE, Flickinger MC (2007) Hydrogen production by photoreactive nanoporous latex coatings of nongrowing Rhodopseudomonas palustris CGA009. Biotechnology Progress 23:124–130
Hillmer P, Gest H (1977) H-2 metabolism in photosynthetic bacterium Rhodopseudomonas capsulata—production and utilization of H-2 by resting cells. J Bacteriol 129:732–739
Melnicki MR, Bianchi L, De Philippis R, Melis A (2008) Hydrogen production during stationary phase in purple photosynthetic bacteria. Int J Hydrogen Energy 33:6525–6534
Eroglu I, Tabanoglu A, Gunduz U, Eroglu E, Yucel M (2008) Hydrogen production by Rhodobacter sphaeroides OU001 in a flat plate solar bioreactor. Int J Hydrogen Energy 33:531–541
Basak N, Das D (2009) Photofermentative hydrogen production using purple non-sulfur bacteria Rhodobacter sphaeroides OU001 in an annular photobioreactor: a case study. Biomass Bioenergy 33:911–919
Kobayashi M, Kobayashi M (1995) Remediation and treatment using anoxygenic phototrophic bacteria. In: Blankenship RE, Madigan MT, Bauer CE (eds) Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers Dordrecht, Netherlands, pp 1269–1282
Thauer RK, Jungermann K, Decker K (1977) Energy conservation in chemotropic anaerobic bacteria. Bacteriol Rev 41:100–180
Franchi E, Tosi C, Scolla G, Della Penna G, Rodriguez F, Pedroni PM (2004) Metabolically engineered Rhodobacter sphaeroides RV strains for improved biohydrogen photoproduction combined with disposal of food wastes. Mar Biotechnol 6:552–565
Hustede E, Steinbuchel A, Schlegel HG (1993) Relationship between the photoproduction of hydrogen and the accumulation of Phb in nonsulfur purple bacteria. Appl Microbiol Biotechnol 39:87–93
Kim MS, Baek JS, Lee JK (2006) Comparison of H-2 accumulation by Rhodobacter sphaeroides KD131 and its uptake hydrogenase and PHB synthase deficient mutant. Int J Hydrogen Energy 31:121–127
Lee IH, Park JY, Kho DH, Kim MS, Lee JK (2002) Reductive effect of H-2 uptake and poly-beta-hydroxybutyrate formation on nitrogenase-mediated H-2 accumulation of Rhodobacter sphaeroides according to light intensity. Appl Microbiol Biotechnol 60:147–153
Zabut B, Ei-Kahlout K, Yucel M, Gunduz U, Turker L, Eroglu I (2006) Hydrogen gas production by combined systems of Rhodobacter sphaeroides OU001 and Halobacterium salinarum in a photobioreactor. Int J Hydrogen Energy 31:1553–1562
Sheng GP, Yu HQ, Yu Z (2005) Extraction of extracellular polymeric substances from the photosynthetic bacterium Rhodopseudomonas acidophila. Appl Microbiol Biotechnol 67:125–130
Watanabe M, Sasaki K, Nakashimada Y, Kakizono T, Noparatnaraporn N, Nishio N (1998) Growth and flocculation of a marine photosynthetic bacterium Rhodovulum sp. Appl Microbiol Biotechnol 50:682–691
Acknowledgments
This research was supported by the Office of Science, Department of Energy, under grants DE-FG02-07ER64495 and DE-FC02-07ER64494. We are indebted to Jackie B. Cooper for her help with analytical equipment and to Rodolfo Perez for valuable discussions and technical help. We also thank Yasar Yilmaz for constructing the light box used in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Submission as a research paper to BioEnergy Research
Rights and permissions
About this article
Cite this article
Yilmaz, L.S., Kontur, W.S., Sanders, A.P. et al. Electron Partitioning During Light- and Nutrient-Powered Hydrogen Production by Rhodobacter sphaeroides . Bioenerg. Res. 3, 55–66 (2010). https://doi.org/10.1007/s12155-009-9072-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-009-9072-8