Abstract
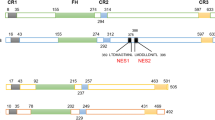
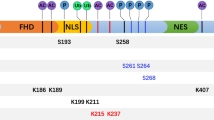
Forkhead box M1 (FoxM1) is an oncogenic transcription factor involved in a wide variety of cellular processes, such as cell cycle progression, proliferation, differentiation, migration, metabolism and DNA damage response. It is overexpressed in many human cancers, especially in breast cancers. Posttranslational modifications are known to play an important role in regulating the expression and transcriptional activity of FoxM1. In this review, we characterize the posttranslational modifications of FoxM1, summarize modifications of FoxM1 by different kinases, explore the relationship between the different sites of modifications and comprehensively describe how posttranslational modifications to regulate the function of FoxM1 by changing protein stability, nucleus localization and transcriptional activity. Additionally, we systematically summarize the roles of FoxM1 in breast cancer occurrence, therapy and drug resistance. The purpose of this paper tries to give a better understanding of the regulatory mechanisms of FoxM1 in cell regulation and highlights potential of a new method for breast cancer therapy by targeting FoxM1.



Similar content being viewed by others
References
Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775(1):92–102. doi:10.1016/j.bbcan.2006.08.006.
Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364(6436):412–20. doi:10.1038/364412a0.
Jiang Y, Liao Y, He H, Xin Q, Tu Z, Kong S, et al. FoxM1 Directs STAT3 expression essential for human endometrial stromal decidualization. Scientific Reports. 2015;5:13735. doi:10.1038/srep13735.
Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6(6):744–50. doi:10.1593/neo.04277.
Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17(3):1626–41.
Wierstra I, Alves J. FOXM1c and Sp1 transactivate the P1 and P2 promoters of human c-myc synergistically. Biochem Biophys Res Commun. 2007;352(1):61–8. doi:10.1016/j.bbrc.2006.10.151.
Wierstra I, Alves J. FOXM1c transactivates the human c-myc promoter directly via the two TATA boxes P1 and P2. FEBS J. 2006;273(20):4645–67. doi:10.1111/j.1742-4658.2006.05468.x.
Wierstra I, Alves J. The central domain of transcription factor FOXM1c directly interacts with itself in vivo and switches from an essential to an inhibitory domain depending on the FOXM1c binding site. Biol Chem. 2007;388(8):805–18. doi:10.1515/bc.2007.094.
Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, et al. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol Cell Biol. 2008;28(9):3076–87. doi:10.1128/mcb.01710-07.
Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25(9):1715–9.
Leung TW, Lin SS, Tsang AC, Tong CS, Ching JC, Leung WY, et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507(1):59–66.
Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN, et al. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215(3):245–52. doi:10.1002/path.2355.
Li SK, Smith DK, Leung WY, Cheung AM, Lam EW, Dimri GP, et al. FoxM1c counteracts oxidative stress-induced senescence and stimulates Bmi-1 expression. J Biol Chem. 2008;283(24):16545–53. doi:10.1074/jbc.M709604200.
Costa RH. FoxM1 dances with mitosis. Nat Cell Biol. 2005;7(2):108–10. doi:10.1038/ncb0205-108.
Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66(4):2153–61. doi:10.1158/0008-5472.can-05-3003.
Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci USA. 2002;99(26):16881–6. doi:10.1073/pnas.252570299.
Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25(24):10875–94. doi:10.1128/mcb.25.24.10875-10894.2005.
Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7(2):126–36. doi:10.1038/ncb1217.
Halasi M, Gartel AL. FOX(M1) news–it is cancer. Mol Cancer Ther. 2013;12(3):245–54. doi:10.1158/1535-7163.mct-12-0712.
Janus JR, Laborde RR, Greenberg AJ, Wang VW, Wei W, Trier A, et al. Linking expression of FOXM1, CEP55 and HELLS to tumorigenesis in oropharyngeal squamous cell carcinoma. Laryngoscope. 2011;121(12):2598–603. doi:10.1002/lary.22379.
Huynh KM, Soh JW, Dash R, Sarkar D, Fisher PB, Kang D. FOXM1 expression mediates growth suppression during terminal differentiation of HO-1 human metastatic melanoma cells. J Cell Physiol. 2011;226(1):194–204. doi:10.1002/jcp.22326.
Teh MT, Gemenetzidis E, Chaplin T, Young BD, Philpott MP. Upregulation of FOXM1 induces genomic instability in human epidermal keratinocytes. Mol Cancer. 2010;9:45. doi:10.1186/1476-4598-9-45.
Nakamura S, Hirano I, Okinaka K, Takemura T, Yokota D, Ono T, et al. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis. 2010;31(11):2012–21. doi:10.1093/carcin/bgq185.
Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M, et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS ONE. 2009;4(3):e4849. doi:10.1371/journal.pone.0004849.
Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67(17):8293–300. doi:10.1158/0008-5472.can-07-1265.
Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM, et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281(35):25167–76. doi:10.1074/jbc.M603906200.
Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66(7):3593–602. doi:10.1158/0008-5472.can-05-2912.
Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66(3):1712–20. doi:10.1158/0008-5472.can-05-3138.
Kalinina OA, Kalinin SA, Polack EW, Mikaelian I, Panda S, Costa RH, et al. Sustained hepatic expression of FoxM1B in transgenic mice has minimal effects on hepatocellular carcinoma development but increases cell proliferation rates in preneoplastic and early neoplastic lesions. Oncogene. 2003;22(40):6266–76. doi:10.1038/sj.onc.1206640.
Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62(16):4773–80.
Mancuso MR, Massarweh SA. Endocrine therapy and strategies to overcome therapeutic resistance in breast cancer. Curr Probl Cancer. 2016. doi:10.1016/j.currproblcancer.2016.09.001.
Saba R, Alsayed A, Zacny JP, Dudek AZ. The role of Forkhead box protein M1 in breast cancer progression and resistance to therapy. Int J Breast Cancer. 2016;2016:9768183. doi:10.1155/2016/9768183.
Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20(5):620–34. doi:10.1016/j.ccr.2011.10.001.
Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24(7):2649–61.
Luscher-Firzlaff JM, Lilischkis R, Luscher B. Regulation of the transcription factor FOXM1c by Cyclin E/CDK2. FEBS Lett. 2006;580(7):1716–22. doi:10.1016/j.febslet.2006.02.021.
Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005;118(Pt 4):795–806. doi:10.1242/jcs.01657.
Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10(9):1076–82. doi:10.1038/ncb1767.
Joshi K, Banasavadi-Siddegowda Y, Mo X, Kim SH, Mao P, Kig C, et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells. 2013;31(6):1051–63. doi:10.1002/stem.1358.
Alvarez-Fernandez M, Halim VA, Aprelia M, Laoukili J, Mohammed S, Medema RH. Protein phosphatase 2A (B55alpha) prevents premature activation of forkhead transcription factor FoxM1 by antagonizing cyclin A/cyclin-dependent kinase-mediated phosphorylation. J Biol Chem. 2011;286(38):33029–36. doi:10.1074/jbc.M111.253724.
Sullivan C, Liu Y, Shen J, Curtis A, Newman C, Hock JM, et al. Novel interactions between FOXM1 and CDC25A regulate the cell cycle. PLoS ONE. 2012;7(12):e51277. doi:10.1371/journal.pone.0051277.
Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27(3):1007–16. doi:10.1128/mcb.01068-06.
Wierstra I, Alves J. Transcription factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biolo Chem. 2006;387(7):949–62. doi:10.1515/bc.2006.119.
Wierstra I. Cyclin D1/Cdk4 increases the transcriptional activity of FOXM1c without phosphorylating FOXM1c. Biochem Biophys Res Commun. 2013;431(4):753–9. doi:10.1016/j.bbrc.2013.01.037.
Duverger O, Chen SX, Lee D, Li T, Chock PB, Morasso MI. SUMOylation of DLX3 by SUMO1 promotes its transcriptional activity. J Cell Biochem. 2011;112(2):445–52. doi:10.1002/jcb.22891.
Abed M, Barry KC, Kenyagin D, Koltun B, Phippen TM, Delrow JJ, et al. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J. 2011;30(7):1289–301. doi:10.1038/emboj.2011.42.
Rytinki MM, Palvimo JJ. SUMOylation modulates the transcription repressor function of RIP140. J Biol Chem. 2008;283(17):11586–95. doi:10.1074/jbc.M709359200.
Gong Z, Brackertz M, Renkawitz R. SUMO modification enhances p66-mediated transcriptional repression of the Mi-2/NuRD complex. Mol Cell Biol. 2006;26(12):4519–28. doi:10.1128/mcb.00409-06.
Yang WH, Heaton JH, Brevig H, Mukherjee S, Iniguez-Lluhi JA, Hammer GD. SUMOylation inhibits SF-1 activity by reducing CDK7-mediated serine 203 phosphorylation. Mol Cell Biol. 2009;29(3):613–25. doi:10.1128/mcb.00295-08.
Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18(22):6455–61. doi:10.1093/emboj/18.22.6455.
Myatt SS, Kongsema M, Man CW, Kelly DJ, Gomes AR, Khongkow P, et al. SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene. 2014;33(34):4316–29. doi:10.1038/onc.2013.546.
Jaiswal N, John R, Chand V, Nag A. Oncogenic Human Papillomavirus 16E7 modulates SUMOylation of FoxM1b. Int J Biochem Cell Biol. 2015;58:28–36. doi:10.1016/j.biocel.2014.11.002.
Schimmel J, Eifler K, Sigurethsson JO, Cuijpers SA, Hendriks IA, Verlaan-de Vries M, et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell. 2014;53(6):1053–66. doi:10.1016/j.molcel.2014.02.001.
Wang CM, Liu R, Wang L, Nascimento L, Brennan VC, Yang WH. SUMOylation of FOXM1B alters its transcriptional activity on regulation of MiR-200 family and JNK1 in MCF7 human breast cancer cells. Int J Mol Sci. 2014;15(6):10233–51. doi:10.3390/ijms150610233.
Karunarathna U, Kongsema M, Zona S, Gong C, Cabrera E, Gomes AR, et al. OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene. 2015. doi:10.1038/onc.2015.208.
Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–9. doi:10.1038/365855a0.
Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, et al. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8(8):869–84.
Whyte P, Williamson NM, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56(1):67–75.
Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5(1):9–15.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi:10.1038/35021093.
Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18(7):830–50. doi:10.1101/gad.1200704.
Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132(4):1420–31. doi:10.1053/j.gastro.2007.01.036.
van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, et al. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol. 2003;163(3):1033–43. doi:10.1016/s0002-9440(10)63463-3.
Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi:10.1186/1471-2407-7-64.
Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388(12):1257–74. doi:10.1515/bc.2007.159.
McMahon G. VEGF receptor signaling in tumor angiogenesis. Oncologist. 2000;5(Suppl 1):3–10.
Graff BA, Bjornaes I, Rofstad EK. Microvascular permeability of human melanoma xenografts to macromolecules: relationships to tumor volumetric growth rate, tumor angiogenesis, and VEGF expression. Microvasc Res. 2001;61(2):187–98. doi:10.1006/mvre.2001.2303.
Zhang Y, Liu J, Wang S, Luo X, Li Y, Lv Z, et al. The DEK oncogene activates VEGF expression and promotes tumor angiogenesis and growth in HIF-1alpha-dependent and -independent manners. Oncotarget. 2016;7(17):23740–56. doi:10.18632/oncotarget.8060.
Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68(21):8733–42. doi:10.1158/0008-5472.can-08-1968.
Park HJ, Gusarova G, Wang Z, Carr JR, Li J, Kim KH, et al. Deregulation of FoxM1b leads to tumour metastasis. EMBO Mole Med. 2011;3(1):21–34. doi:10.1002/emmm.201000107.
Wang X, Bhattacharyya D, Dennewitz MB, Kalinichenko VV, Zhou Y, Lepe R, et al. Rapid hepatocyte nuclear translocation of the Forkhead Box M1B (FoxM1B) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expr. 2003;11(3–4):149–62.
Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26(42):6212–9. doi:10.1038/sj.onc.1210443.
Yang C, Chen H, Tan G, Gao W, Cheng L, Jiang X, et al. FOXM1 promotes the epithelial to mesenchymal transition by stimulating the transcription of Slug in human breast cancer. Cancer Lett. 2013;340(1):104–12. doi:10.1016/j.canlet.2013.07.004.
Bektas N, Haaf A, Veeck J, Wild PJ, Luscher-Firzlaff J, Hartmann A, et al. Tight correlation between expression of the Forkhead transcription factor FOXM1 and HER2 in human breast cancer. BMC Cancer. 2008;8:42. doi:10.1186/1471-2407-8-42.
Bergamaschi A, Madak-Erdogan Z, Kim YJ, Choi YL, Lu H, Katzenellenbogen BS. The forkhead transcription factor FOXM1 promotes endocrine resistance and invasiveness in estrogen receptor-positive breast cancer by expansion of stem-like cancer cells. Breast Cancer Res BCR. 2014;16(5):436. doi:10.1186/s13058-014-0436-4.
Ahn H, Sim J. Increased expression of forkhead box M1 is associated with aggressive phenotype and poor prognosis in estrogen receptor-positive breast cancer. J Korean Med Sci. 2015;30(4):390–7. doi:10.3346/jkms.2015.30.4.390.
Horimoto Y, Hartman J, Millour J, Pollock S, Olmos Y, Ho KK, et al. ERbeta1 represses FOXM1 expression through targeting ERalpha to control cell proliferation in breast cancer. Am J Pathol. 2011;179(3):1148–56. doi:10.1016/j.ajpath.2011.05.052.
Francis RE, Myatt SS, Krol J, Hartman J, Peck B, McGovern UB, et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol. 2009;35(1):57–68.
Hamurcu Z, Ashour A, Kahraman N, Ozpolat B. FOXM1 regulates expression of eukaryotic elongation factor 2 kinase and promotes proliferation, invasion and tumorgenesis of human triple negative breast cancer cells. Oncotarget. 2016;7(13):16619–35. doi:10.18632/oncotarget.7672.
Park YY, Jung SY, Jennings NB, Rodriguez-Aguayo C, Peng G, Lee SR, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis. 2012;33(10):1843–53. doi:10.1093/carcin/bgs167.
Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM, et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010;29(20):2983–95. doi:10.1038/onc.2010.47.
Nestal de Moraes G, Delbue D, Silva KL, Robaina MC, Khongkow P, Gomes AR, et al. FOXM1 targets XIAP and Survivin to modulate breast cancer survival and chemoresistance. Cell Signal. 2015;27(12):2496–505. doi:10.1016/j.cellsig.2015.09.013.
Khongkow P, Gomes AR, Gong C, Man EP, Tsang JW, Zhao F, et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35(8):990–1002. doi:10.1038/onc.2015.152.
Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mole Cancer Res MCR. 2010;8(1):24–34. doi:10.1158/1541-7786.mcr-09-0432.
McGovern UB, Francis RE, Peck B, Guest SK, Wang J, Myatt SS, et al. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol Cancer Ther. 2009;8(3):582–91. doi:10.1158/1535-7163.mct-08-0805.
Millour J, de Olano N, Horimoto Y, Monteiro LJ, Langer JK, Aligue R, et al. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol Cancer Ther. 2011;10(6):1046–58. doi:10.1158/1535-7163.mct-11-0024.
Monteiro LJ, Khongkow P, Kongsema M, Morris JR, Man C, Weekes D, et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene. 2013;32(39):4634–45. doi:10.1038/onc.2012.491.
Arora R, Yates C, Gary BD, McClellan S, Tan M, Xi Y, et al. Panepoxydone targets NF-kB and FOXM1 to inhibit proliferation, induce apoptosis and reverse epithelial to mesenchymal transition in breast cancer. PLoS ONE. 2014;9(6):e98370. doi:10.1371/journal.pone.0098370.
Ahmad A, Ali S, Wang Z, Ali AS, Sethi S, Sakr WA, et al. 3,3′-Diindolylmethane enhances taxotere-induced growth inhibition of breast cancer cells through downregulation of FoxM1. Int J Cancer. 2011;129(7):1781–91. doi:10.1002/ijc.25839.
Yu G, Zhou A, Xue J, Huang C, Zhang X, Kang SH, et al. FoxM1 promotes breast tumorigenesis by activating PDGF-A and forming a positive feedback loop with the PDGF/AKT signaling pathway. Oncotarget. 2015;6(13):11281–94. doi:10.18632/oncotarget.3596.
Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008;7(7):2022–32. doi:10.1158/1535-7163.MCT-08-0188.
Wang JS, Ren TN, Xi T. Ursolic acid induces apoptosis by suppressing the expression of FoxM1 in MCF-7 human breast cancer cells. Med Oncol. 2012;29(1):10–5. doi:10.1007/s12032-010-9777-8.
Acknowledgements
This work was partly supported by the National Nature Science Foundation of China (Grant Nos 21405138 and 81273111), the Scientific Projects of Zhejiang Province (2015C33148 and 2015C37117), the Foundations of Innovative Research Team of Educational Commission of Zhejiang Province (T200907), Ningbo Scientific Innovation Team for Environmental Hazardous Factor Control and Prevention (2016C51001) and KC Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
None.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Song, X., Fiati Kenston, S.S., Zhao, J. et al. Roles of FoxM1 in cell regulation and breast cancer targeting therapy. Med Oncol 34, 41 (2017). https://doi.org/10.1007/s12032-017-0888-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-017-0888-3