Abstract
Gastropulmonary route of infection was considered to be an important mechanism of ventilator-associated pneumonia (VAP). However there is little evidence to support this assumption. Moreover, the prevalence of microaspiration in elderly ventilated patients was not well understood. To confirm gastropulmonary infection route and investigate the prevalence of microaspiration in elderly ventilated patients using genome macrorestriction-pulsed field gel electrophoresis (GM-PFGE). Patients over 60 years old, expected to receive mechanical ventilation longer than 48 h, were prospectively enrolled from October 2009 to January 2012. Clinical data were collected and recorded until they died, developed pneumonia, or were extubated. Samples from gastric fluid, subglottic secretion and lower respiratory tract (LRT) were collected during the follow-up for microbiological examination. To evaluate the homogeneity, GM-PFGE was performed on strains responsible for VAP that had the same biochemical phenotype as those isolated from gastric juice and subglottic secretions sequentially. Among 44 VAP patients, 76 strains were isolated from LRT and considered responsible for VAP. Twenty-two isolates had the same biochemical phenotype with the corresponding gastric isolates. The homology was further confirmed using GM-PFGE in 12 episodes of VAP. Nearly 30 % of VAPs were caused by microaspiration based on the analysis of bacterial phenotype or GM-PFGE. In addition, 58.3 % patients with gastric colonization developed VAP, especially late-onset VAP (LOP). Gastropulmonary infection route exists in VAP especially LOP in elderly ventilated patients. It is one of the important mechanisms in the development of VAP.
Similar content being viewed by others
Introduction
Ventilator-associated pneumonia (VAP) is a nosocomial pneumonia that typically occurs in patients receiving mechanical ventilation (MV) for longer than 48 h. The incidence rate varies from 6 to 52 % and the attributable mortality ranges from 24 to 76 % in critically ill patients [1, 2]. Patients over 60 years old are more likely to suffer from VAP [2]. The colonization of stomach, oropharynx, and trachea by potential microaspiration may play an important role in the onset of VAP in critically ill intubated patients. Measures aiming to prevent aspiration in intubated patients could be helpful in preventing VAP and improving patient outcomes [3, 4]. However, there are no sufficient microbiological evidences to support the gastric colonization and gastropulmonary route of infection in the pathogenesis of VAP. Several studies tried to use Technetium 99 labeled enteral feeding, blue methylene or quantitative pepsin measurement in tracheal aspirates to verify the microaspiration in critically ill patients [4–7]. However, radioactivity or difficulty in implementation limits the application of these techniques. The prevalence of microaspiration in the airway in elderly ventilated patients is also unknown. With the aging of the society, more attentions should be paid to the elderly patients. More useful measures to prevent VAP especially in elderly patients should be taken into account. The aims of this study are: (1) to further verify gastropulmonary route of infection in the pathogenesis of VAP in elderly ventilated patients by testing homogeneity of the microorganisms isolated from gastric juice, subglottic secretions, and endotracheal aspirates sequentially with genome macrorestriction-pulsed field gel electrophoresis (GM-PFGE) and to develop an easy and sensitive diagnosis tool for microaspiration; (2) to investigate the prevalence of microaspiration in elderly ventilated patients in order to develop effective prevention strategies for VAP.
Methods and Materials
Study Design
This study was conducted at the Hospital from October 2009 to January 2012. Inclusion criteria were patients expecting mechanical ventilation for more than 48 h. Because of the recession of laryngopharyngeal sense, microaspiration is more likely to occur in older patient than in the younger ones. To better prove the role of microaspiration and gastropulmonary route in VAP, patients younger than 60 years old were excluded from the study. Patients were also excluded if they died within 48 h, or the duration of MV was shorter than 48 h or had pre-existing pneumonia when MV began. Patients were followed until death, incidence of pneumonia, or extubation. For strains responsible for VAP that were identical to those isolated from gastric juices, subglottic secretions, and LRT in biochemical phenotype, chromosomal DNA analysis was performed with GM-PFGE. All patients received prokinetic drug as routine during the study. Hospital’s Human Subject Research Committee approved the study prior to its initiation. Informed consent was also obtained from each patient or their legally authorized representatives.
A clinically suspected VAP was defined as the presence of a new persistent or progressive infiltration on chest X-ray and at least 3 of the four following criteria [8]: (1) rectal temperature >38.0 °C or <35.5 °C; (2) Blood leukocytosis (>10 × 103/mm3) and/or left shift of leucopenia (<3 × 103/mm3); (3) >10 leukocytes per high-power field in Gram stain of tracheal aspirate; (4) tracheal aspirate semi-quantitative culture >(+). When VAP is suspected, bronchoscopy with protected specimen brush (PSB) was performed. The diagnosis of VAP was established on the basis of positive quantitative cultures from PSB [cutoff point ≥103 colony-forming units (cfu)/ml] [9]. Based on the time of onset, VAP can be divided into 2 types: early- and late-onset. Early-onset VAP (EOP) occurs between 48 and 96 h after intubation, while late-onset VAP (LOP) occurs later than 96 h after intubation.
Collection and Measurement of Key Variables
Descriptive data were collected, including demographics, co-morbidities, severity of illness as determined by scores on the Acute Physiology, Age, and Chronic Health Evaluation (APACHE) II, and ICU outcomes. Demographic data included gender, age, clinical settings, and reasons for administration.
Gram stain and culture of the subglottic secretions, endotracheal aspirates, and gastric juice were performed every day. Chest X-ray was taken prior to MV, on the 1st, 7th, 15th day of MV or whenever nosocomial pneumonia was suspected. On the 4th day of MV, 2 ml methylene blue diluted with 20 ml normal solution was administered to the patient through the nasogastric tube.
All the data were recorded on specific case report forms.
Chromosomal DNA Analysis by GM-PFGE
Chromosomal DNA analysis with GM-PFGE was performed for isolates that were collected from the lower respiratory tract (LRT), responsible for VAP. These isolates were the identical pathogens in bacterial culture (biochemical phenotype) to those isolated from gastric juices and subglottic secretions.
All specimens were inoculated onto blood agar immediately after sampling. One colony was selected and cultured on Luria-Bertain substrate (LB) at 37 °C for 24–28 h, and then centrifuged, followed by resuspension in TE25S buffer (saccharose 0.3 M, 0 25 mM EDTA- Tris–HCl, pH 8.0). The bacterial concentration was then adjusted to OD550 equal to 1.0. The suspensions were mixed with equal volume of 2 % low melting point agarose and allowed to solidify in 100 μl molds. Plugs were incubated in 1 ml of TE25S solution with 1–2 mg/ml of lysozyme for 2–4 h at 37 °C and then incubated in 1 % NDS solution [EDTA (pH 8.0) 0.5 mM, Sodium Lauroyl Sarcosine 1.0 % (AMRESCO, Shanghai bioengineering Ltd.)] supplemented with 200 μg/ml of proteinase K (AMRESCO) for 24 h at 50 °C. After washing with 1 ml TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]), the samples were digested with 30–60 U of Sma I, Xbal (Fermentas life science, Shanghai bioengineering Ltd.) or Spe I (New England Biolab, NEB, Shanghai, Ltd.) for 2 h according to the manufacturer's recommendations. Fragments of DNA were separated using PFGE in a 1 % agarose gel (AMRESCO, Shanghai bioengineering Ltd.) in 0.5× TBE buffer [diluted from 5× TBE (Tris alkali 54 g, boracic acid 27.5 g, 0.5 M EDTA, pH 8.0)] at 14 °C using the Bio-Rad CHEF-DR XA system. Then samples and the DNA marker (lambda ladder, ProMega Ltd.) were electrophoresised at 6 V/cm for 20 h with pulse time ranging from 5 to 45 s depending on the different strains. After electrophoresis, gels were stained with 0.5 μg/ml ethidium bromide (EB) for 1 h, washed, and photographed with Bio-rad visualization system. Strains were differentiated according to the criteria of Tenover et al. [10].
Statistical Analysis
Statistical analysis was performed with Statistical Package for the Social Sciences, version 18.0 for Windows (SPSS Inc., Chicago, IL, USA). Continuous variables were summarized as mean ± standard deviation (SD) and categorical variables were summarized as percentage of the study population. Categorical variables were compared using Pearson χ 2 test. All tests were two-sided, and P value <0.05 was considered to be statistically significant.
Results
Patients Characteristics and Incidence of VAP
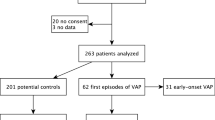
Over a period of 15 months, 112 eligible patients were screened. Eighty-six patients met the inclusion criteria, including 53 males and 33 females. The average age was 70.5 ± 8.1 years, ranging from 60 to 92 years. Sixty-five patients (75.6 %) were from surgical ICU, the rest from respiratory ICU, cardiac ICU, medical ICU, and emergency room. Forty-four patients (51.2 %) developed VAP during the study. The demographic and clinical characteristic of the study population were shown in Table 1.
The Relationship Between VAP and Gastric Colonization
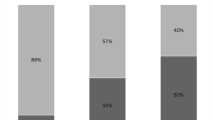
Of the 86 study patients, there were 60 with positive cultures of gastric samples, and 26 with negative culture. Among these sixty patients, 58.3 % (35 patients) developed VAP. In comparison, only 9 out of 26 patients (23.1 %) with no bacterial colonization in gastric juice developed VAP (P = 0.043) (Table 2).
Pathogens Isolated from Different Body Sites and Aspiration of Oropharyngeal Secretions
Seventy-six different strains were isolated from LRT and regarded as the causative pathogen in all 44 episodes of VAP. Of all the 76 stains isolated from LRT, 22 (18 isolated from episode of late-onset VAP) were proved to have the identical biochemical phenotype with those isolated from the gastric fluid. Late-onset VAP was defined as VAP developed after more than 5 days of MV. This suggested that about 28.9 % (22/76) causative pathogens for VAP might have come from gastric fluid. Table 3 summarized the pathogens isolated from LRT and gastric fluid. Eighteen cases received methylene blue through nasogastric tube. Methylene blue was invisible but could be detected in oropharyngeal secretions in all 18 patients under the microscope.
Gastropulmonary Route of Infection Analyzed by Chromosomal DNA Analysis with GM-PFGE
Biochemical typing and antibiotic susceptibility test were done for all isolated pathogens. Thirty-five phenotypically identical strains were isolated from gastric fluid, subglottic secretions, and LRT in 15 episodes of VAP. GM-PFGE was performed to confirm the homology. The result showed a homology of strains in 12 episodes of VAP (12/44, 27.3 %). All these strains were isolated from gastric samples or LRT except one strain of Streptococcus pneumoniae isolated from subglottic secretions. Figure 1 showed the GM-PFGE results of these 35 strains. It is notable that the fragments of Acinetobacter baumannii or methicillin-resistant Staphylococcus aureus (MRSA) isolated from different patients didn’t show any significant difference (Fig. 1d).
Pulsed-field gel electrophoresis (PFGE) of strains with identical biochemical phenotypes that were sequentially isolated from gastric juice, lower respiratory tract (LRT), and subglottic secretions. a 15 Pseudomonas aeruginosa stains. Lane 1 phage lambda DNA (Marker); lanes 2 gastric samples, lane 3 LRT of the same patients, showing that there were more than 3 different fragments between them; lanes 4, 5 and lanes 15, 16 were isolates from the gastric samples and LRT of 2 patients, respectively. Lanes 6–8, lanes 9–11, and lanes 12–14 were isolates from the gastric samples, subglottic samples and LRT of 3 patients, respectively. For these 5 patients, PFGE analysis confirmed that the pathogens from different sites were identical. b 9 Enterobacteriaceae strains. Lane 1 phage lambda DNA (Marker); lanes 2, 3 Escherichia coli from gastric samples and LRT of the same patient. Lanes 4, 5 Klebsiella pneumoniae from gastric samples and LRT of the same patient. Lanes 6, 7 Enterobacter cloacae from gastric samples and LRT of the same patient. Lanes 8–10 E. cloacae from gastric samples, LRT and subglottic samples of the same patients. All strains of E. cloacae isolated from the same patient were identical. c 2 Streptococcus pneumoniae strains. Lane 1 phage lambda DNA (Marker). One different fragment between 2 strains was showed. d 4 Staphylococcus aureus stains and 5 Acinetobacter baumannii strains. Lane 1 phage lambda DNA (Marker); lanes 2, 3 and lanes 4, 5 Staphylococcus aureus isolated from the gastric samples and LRT of 2 patients, respectively. Lanes 6, 7 A. baumannii isolated from gastric sample and LRT of the same patient. Lanes 8–10 A. baumannii from gastric samples, LRT and subglottic samples of another patient
Discussion
The role of the gastric reservoir in the pathogenesis of VAP is still controversial [11]. Aspiration of human gastric juice might associate with tracheobronchitis and VAP. The incidence of aspiration varies greatly from 11.3 to 88 % of intubated patients [12, 13]. In this study, we found that about 28.9 % causative pathogens for VAP had the identical phenotype with those isolated from gastric fluids. This result is consistent with our previous adult cohort study [14]. Moreover, we found that VAP developed more frequently in patients with positive gastric content culture. These results indicate microorganisms colonizing in the gastrointestinal tract and microaspiration may be very important factors for the development of VAP. This was further confirmed by chromosomal DNA analysis using GM-PFGE. In our study we found that the bacterial strains that caused VAP were identical to those previously colonized in the stomach and the incidence of microaspiration was about 27.3 % using GM-PFGE analysis. Similar results were also reported by Garrouste-Orgeas et al. [15]. Several other studies suggested that administration of prokinetic drugs combined with semi-recumbent body positioning and/or continuous subglottic secretions drainage could be helpful in reducing VAP, especially in the elderly patients [16–18].
How to detect the microaspiration easily and accurately remains challenging. Methylene blue is probably the most frequently used marker for aspiration in animal and human studies. However, it is impossible to quantitatively determine microaspiration, and fiberoptic bronchoscopy is required to diagnose microaspiration with blue dye. Pepsin measurement in tracheal aspirates is accurate and easy to use in diagnosing microaspiration of gastric contents in critically ill patients. However, pepsin should be detected rapidly after aspiration [4, 12]. In this study bacterial culture and/or GM-PFGE were used to confirm the microaspiration in ventilated elderly patients. GM-PFGE is considered the gold standard for detecting the pathogens’ homogeneity. The results of bacterial culture were highly consistent with the result of GM-PFGE in our study. As we know, bacterial culture is easy to perform in clinic. And it is also beneficial for choosing appropriate and effective antibiotics. The disadvantage of bacterial culture is that it may take 72 h to get the results, which might delay the diagnosis. Gram strain is a rather quicker and easier method than bacterial culture. But further study is needed to determine whether gram strain could become an alternative choice. Bacterial culture or/and Gram stain of the secretions from different parts of the body could help detect the microaspiration in patient with MV timely. Method to further prevent VAP should be studied based on this pathogenesis.
Unfortunately, we noticed that some Acinetobacter baumannii and Staphylococcus aureus strains had identical molecular type in different patients using GM-PFGE analysis. This demonstrated the occurrence of cross-infection in ICU. Prevention and control of hospital infection should be strengthened in the future.
Our study has some limitations. The cases that met the study criteria were mainly postoperative surgical patients from the same hospital. Multicenter research should be considered to illustrate the gastropulmonary route of infection and prevalence of microaspiration for VAP in elderly patients in the future. We also noticed that the incidence of VAP was nearly 2 times high as that reported by Kollef et al. [19] and Liberati et al. [20] recently. According to the epidemiological study and continuous monitoring of ventilation-associated pneumonia in our hospital, the incidence of VAP was as high as 51.2 % in surgical ICU from December 1999 to February 2001 [21]. The possible explanation is that the subjects enrolled in this study were elderly patients who were at a higher risk of developing pneumonia. Another underlying reason is that the clinical practice of infection control such as strict hand hygiene was not well recognized and implemented during the study period.
In conclusion, this study demonstrated the existence of gastropulmonary infection route for VAP, especially the late-onset VAP. It may play an important role in the development of VAP. The results of bacterial phenotyping were consistent with GM-PFGE. Further research of VAP prevention based on this pathogenesis is needed.
References
Joseph, N. M., Sistla, S., Dutta, T. K., Badhe, A. S., & Parija, S. C. (2010). Ventilator-associated pneumonia: a review. European Journal of Internal Medicine, 21(5), 360–368. doi:10.1016/j.ejim.2010.07.006.
Werarak, P., Kiratisin, P., & Thamlikitkul, V. (2010). Hospital-acquired pneumonia and ventilator-associated pneumonia in adults at Siriraj Hospital: Etiology, clinical outcomes, and impact of antimicrobial resistance. Journal of the Medical Association of Thailand, 93(Suppl 1), S126–S138.
Kamat, P., Favaloro-Sabatier, J., Rogers, K., & Stockwell, J. A. (2008). Use of methylene blue spectrophotometry to detect subclinical aspiration in enterally fed intubated pediatric patients. Pediatric Critical Care Medicine, 9(3), 299–303. doi:10.1097/PCC.0b013e318172d500.
Nseir, S., Zerimech, F., Jaillette, E., Artru, F., & Balduyck, M. (2011). Microaspiration in intubated critically ill patients: diagnosis and prevention. Infectious Disorders: Drug Targets, 11(4), 413–423.
Gadani, H., Vyas, A., & Kar, A. K. (2010). A study of ventilator-associated pneumonia: Incidence, outcome, risk factors and measures to be taken for prevention. Indian Journal of Anaesthesia, 54(6), 535–540. doi:10.4103/0019-5049.72643.
Muscedere, J., Rewa, O., McKechnie, K., Jiang, X., Laporta, D., & Heyland, D. K. (2011). Subglottic secretion drainage for the prevention of ventilator-associated pneumonia: A systematic review and meta-analysis. Critical Care Medicine, 39(8), 1985–1991. doi:10.1097/CCM.0b013e318218a4d9.
Wang, F., Bo, L., Tang, L., Lou, J., Wu, Y., Chen, F., et al. (2012). Subglottic secretion drainage for preventing ventilator-associated pneumonia: an updated meta-analysis of randomized controlled trials. Journal of Trauma and Acute Care Surgery, 72(5), 1276–1285. doi:10.1097/TA.0b013e318247cd33.
Smulders, K., van der Hoeven, H., Weers-Pothoff, I., & Vandenbroucke-Grauls, C. (2002). A randomized clinical trial of intermittent subglottic secretion drainage in patients receiving mechanical ventilation. Chest, 121(3), 858–862.
Bergmans, D. C., Bonten, M. J., Gaillard, C. A., Paling, J. C., van der Geest, S., van Tiel, F. H., et al. (2001). Prevention of ventilator-associated pneumonia by oral decontamination: a prospective, randomized, double-blind, placebo-controlled study. American Journal of Respiratory and Critical Care Medicine, 164(3), 382–388.
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. Journal of Clinical Microbiology, 33(9), 2233–2239.
Augustyn, B. (2007). Ventilator-associated pneumonia: risk factors and prevention. Critical Care Nurse, 27(4), 32–36, 38–39; quiz 40.
Metheny, N. A., Clouse, R. E., Chang, Y. H., Stewart, B. J., Oliver, D. A., & Kollef, M. H. (2006). Tracheobronchial aspiration of gastric contents in critically ill tube-fed patients: Frequency, outcomes, and risk factors. Critical Care Medicine, 34(4), 1007–1015. doi:10.1097/01.CCM.0000206106.65220.59.
Miller, C. D., Rebuck, J. A., Ahern, J. W., & Rogers, F. B. (2005). Daily evaluation of macroaspiration in the critically ill post-trauma patient. Current Surgery, 62(5), 504–508. doi:10.1016/j.cursur.2005.03.003.
Li, H. Y., He, L. X., Hu, B. J., Wang, B. Q., Zhang, X. Y., Chen, X. H., et al. (2004). The impact of gastric colonization on the pathogenesis of ventilator-associated pneumonia. Zhonghua Nei Ke Za Zhi, 43(2), 112–116.
Garrouste-Orgeas, M., Chevret, S., Arlet, G., Marie, O., Rouveau, M., Popoff, N., et al. (1997). Oropharyngeal or gastric colonization and nosocomial pneumonia in adult intensive care unit patients. A prospective study based on genomic DNA analysis. American Journal of Respiratory and Critical Care Medicine, 156(5), 1647–1655.
Bouza, E., Perez, M. J., Munoz, P., Rincon, C., Barrio, J. M., & Hortal, J. (2008). Continuous aspiration of subglottic secretions in the prevention of ventilator-associated pneumonia in the postoperative period of major heart surgery. Chest, 134(5), 938–946. doi:10.1378/chest.08-0103.
Mizock, B. A. (2007). Risk of aspiration in patients on enteral nutrition: frequency, relevance, relation to pneumonia, risk factors, and strategies for risk reduction. Current Gastroenterology Reports, 9(4), 338–344.
Ramirez, P., Bassi, G. L., & Torres, A. (2012). Measures to prevent nosocomial infections during mechanical ventilation. Current Opinion in Critical Care, 18(1), 86–92. doi:10.1097/MCC.0b013e32834ef3ff.
Kollef, K. E., Schramm, G. E., Wills, A. R., Reichley, R. M., Micek, S. T., & Kollef, M. H. (2008). Predictors of 30-day mortality and hospital costs in patients with ventilator-associated pneumonia attributed to potentially antibiotic-resistant gram-negative bacteria. Chest, 134(2), 281–287. doi:10.1378/chest.08-1116.
Liberati, A., D’Amico, R., Pifferi, S., Torri, V., Brazzi, L., & Parmelli, E. (2009). Antibiotic prophylaxis to reduce respiratory tract infections and mortality in adults receiving intensive care. Cochrane Database of Systematic Reviews, 4, CD000022. doi:10.1002/14651858.CD000022.pub3.
Liu, Q. H., He, L. X., Hu, B. J., Li, H. Y., Chen, X. H., Gao, X. D., et al. (2006). Comprehensive prevention and pathogenesis of ventilator-associated pneumonia in elderly patients: a prospective, randomized, case-control clinical trial. Zhonghua Nei Ke Za Zhi, 45(9), 717–720.
Acknowledgments
This study was supported by Project of Tenth Five-Year Plan from National Department of Technology (No. 2001BA702B06), National Basic Research Program of China (973 Program) (No. 2007CB513000), Shandong Province Outstanding Young Scientists Research Award Fund (No. BS2009YY043), and Jinan Municipal Science and Technology Bureau of Jinan City, the development of science and technology in 2010, the fourth installment plan project (university institutes independent innovation plan) (No. 201004058). The authors acknowledge the surgical ICU staff for their help in recruiting the patients and Dr. Ying-ming Zhu from Shanghai Scientific Research Institute for giving advice on PFGE techniques. Additionally we thank all technicians in the microbiological laboratory in Zhongshan hospital.
Conflict of interest
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Qing-hua Liu and Jing Zhang have contributed equally to this article.
Rights and permissions
About this article
Cite this article
Liu, Qh., Zhang, J., Lin, Dj. et al. Gastropulmonary Route of Infection and the Prevalence of Microaspiration in the Elderly Patients with Ventilator-Associated Pneumonia Verified by Molecular Microbiology-GM-PFGE. Cell Biochem Biophys 71, 1457–1462 (2015). https://doi.org/10.1007/s12013-014-0368-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-014-0368-8