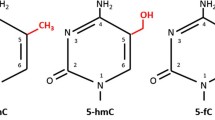
Abstract
The profile of allergic disease worldwide continues to change as the number of severe IgE-mediated allergies increases. This phenomenon is thought to reflect the outcome of combined genetic/environmental/developmental/stochastic effects on immune development, but understanding this remains a challenge. Epigenetic disruption at key immune genes during development has been proposed as a potential explanation for how environmental exposures may alter immune cell development and function. This represents an emerging area of research with the potential to yield new understanding of how disease risk is modified. Here, we examine recent developments in this field that are defining new epigenetic paradigms of allergic disease.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Poulos LM, Waters A-M, Correll PK, Loblay RH, Marks GB. Trends in hospitalizations for anaphylaxis, angioedema, and urticaria in Australia, 1993–1994 to 2004–2005. Journal of allergy and clinical immunology. 2007;120:878–84.
Tang MLK, Osborne N, Allen K. Epidemiology of anaphylaxis. Curr Opin Allergy Clin Immunol. 2009;9:351–6.
Lin RY, Anderson AS, Shah SN, Nurruzzaman F. Increasing anaphylaxis hospitalizations in the first 2 decades of life: New York State, 1990–2006. Ann Allergy Asthma Immunol. 2008;101:387–93.
Lee JK, Vadas P. Anaphylaxis: mechanisms and management. Clin Exp Allergy. 2011;41:923–38.
Allen KJ, Koplin JJ. The epidemiology of IgE-mediated food allergy and anaphylaxis. Immunol Allergy Clin North Am. 2012;32:35–50.
Koplin JJ, Martin PE, Allen KJ. An update on epidemiology of anaphylaxis in children and adults. Curr Opin Allergy Clin Immunol. 2011;11:492–6.
Cianferoni A, Muraro A. Food-induced anaphylaxis. Immunol Allergy Clin North Am. 2012;32:165–95.
Prescott S, Allen KJ. Food allergy: Riding the second wave of the allergy epidemic. Pediatr Allergy Immunol. 2011;22:155–60.
Mullins RJ, Dear KBG, Tang MLK. Characteristics of childhood peanut allergy in the Australian Capital Territory, 1995 to 2007. J Allergy Clin Immunol. 2009;123:689–93.
Fazekas de St Groth B. Regulatory T-cell abnormalities and the global epidemic of immuno-inflammatory disease. Immunol Cell Biol. 2012;90:256–9.
Kondilis-Mangum HD, Wade PA. Epigenetics and the adaptive immune response. Mol Aspects Med. 2012. doi:10.1016/j.mam.2012.06.008.
Suarez-Alvarez B, Rodriguez RM, Fraga MF, López-Larrea C. DNA methylation: a promising landscape for immune system-related diseases. Trends Genet 2012, In press.
Palm NW, Rosenstein RK, Medzhitov R. Allergic host defenses. Nature. 2012;484:465–72.
North ML. Ellis AK The role of epigenetics in the developmental origins of allergic disease. Ann Allergy Asthma Immunol. 2011;106:355–61.
Bell C, Finer S, Lindgren C, Wilson G. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS ONE. 2010;5:1–12.
Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA Methyltransferase 1 Is Essential for and Uniquely Regulates Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 2009;5:442–9.
Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–7.
Broeske A-M, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–U69.
White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO- T cells. J Immunol. 2002;168:2820–7.
White GP, Hollams EM, Yerkovich ST, Bosco A, Holt BJ, Bassami MR, Kusel M, Sly PD, Holt PG. CpG methylation patterns in the IFN gamma; promoter in naive T cells: Variations during Th1 and Th2 differentiation and between atopics and non-atopics. Pediatr Allergy Immunol. 2006;17:557–64.
Porrás A, Kozar S, Russanova V, et al. Developmental and epigenetic regulation of the human TLR3 gene. Mol Immunol. 2008;46:27–36.
Goriely S, Van Lint C, Dadkhah R, Libin M, De Wit D, Demonte D, Willems F, Goldman M. A defect in nucleosome remodeling prevents IL-12(p35) gene transcription in neonatal dendritic cells. J Exp Med. 2004;199:1011–6.
Kim SY, Romero R, Tarca AL, et al. Methylome of Fetal and Maternal Monocytes and Macrophages at the Feto-Maternal Interface. Am J Reprod Immunol. 2012. doi:10.1111/j.1600-0897.2012.01108.x.
Susan L Prescott, Annett Osei-Kumah, Tara Richman, Megan Hodder, David Martino, Janet A Dunstan, Meri Tulic, Boris Novakovic, Safferry R, Clifton VL Differential placental FOXP3 and Th2 gene expression with sex-specific differences in infant allergy. 2011, 128: 886–7
Slaats GGG, Reinius LE, Alm J, Kere J, Scheynius A, Joerink M. DNA methylation levels within the CD14 promoter region are lower in placentas of mothers living on a farm. Allergy. 2012;67:895–903.
Smith M, Tourigny MR, Noakes P, Thornton CA, Tulic MK, Prescott SL. Children with egg allergy have evidence of reduced neonatal CD4 + CD25 + CD127lo/- regulatory T cell function. Journal of allergy and clinical immunology. 2008;121:1460–6.
Prescott SL, Noakes P, Chow BWY, Breckler L, Thornton CA, Hollams EM, Ali M, van den Biggelaar AHJ, Tulic MK. Presymptomatic differences in Toll-like receptor function in infants who have allergy. J Allergy Clin Immunol. 2008;122:391–9. 399.e1–5.
• Hinz D, Bauer M, Roder S, et al. Cord blood Tregs with stable FOXP3 expression are influenced by prenatal environment and associated with atopic dermatitis at the age of one year. Allergy. 2012;67:380–9. Of importance: This study demonstrates the utility of DNA methylation marks as surrogate markers of Treg cell numbers. The authors use a unique methodology to report an association between prenatal exposures, variations in Treg DNA methylation and allergic outcomes in infancy.
Janson PCJ, Winerdal ME, Winqvist O. At the crossroads of T helper lineage commitment-Epigenetics points the way. Bba-Gen Subjects. 2009;1790:906–19.
Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–67.
Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C. Hierarchical IL-5 expression defines a subpopulation of highly differentiated human Th2 cells. J Immunol. 2011;187:3111–20.
Cohen CJ, Crome SQ, Macdonald KG, Dai EL, Mager DL, Levings MK. Human Th1 and th17 cells exhibit epigenetic stability at signature cytokine and transcription factor Loci. J Immunol. 2011;187:5615–26.
Nistala K, Adams S, Cambrook H, Ursu S, Olivito B, de Jager W, Evans JG, Cimaz R, Bajaj-Elliott M, Wedderburn LR. Th17 plasticity in human autoimmune arthritis is driven by the inflammatory environment. Proc Natl Acad Sci U S A. 2010;107:14751–6.
• Swamy RS, Reshamwala N, Hunter T, Vissamsetti S, Santos CB, Baroody FM, Hwang PH, Hoyte EG, Garcia MA, Nadeau KC. Epigenetic modifications and improved regulatory T-cell function in subjects undergoing dual sublingual immunotherapy. J Allergy Clin Immunol. 2012;130:215–24. Of importance. This study demonstrates some clear epigenetic changes in key immune cell populations in response to allergen therapy. These epigenetic events are clearly associated with altered immune outcomes.
Brand S, Kesper DA, Teich R, Kilic-Niebergall E, Pinkenburg O, Bothur E, Lohoff M, Garn H, Pfefferle PI, Renz H. DNA methylation of T(H)1/T(H)2 cytokine genes affects sensitization and progress of experimental asthma. J Allergy Clin Immunol. 2012. doi:10.1016/j.jaci.2011.12.963.
Allan RS, Zueva E, Cammas F, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature. 2012. doi:10.1038/nature11173.
Niedzwiecki M, Zhu H, Corson L, Grunig G, Factor PH, Chu S, Jiang H, Miller RL. Prenatal exposure to allergen, DNA methylation, and allergy in grandoffspring mice. Allergy. 2012. doi:10.1111/j.1398-9995.2012.02841.x.
Xue X, Feng T, Yao S, Wolf KJ, Liu C-G, Liu X, Elson CO, Cong Y. Microbiota Downregulates Dendritic Cell Expression of miR-10a, Which Targets IL-12/IL-23p40. J Immunol. 2011;187:5879–86.
Mullins RJ, Camargo CA. Latitude, sunlight, vitamin D, and childhood food allergy/anaphylaxis. Curr Allergy Asthma Rep. 2012;12:64–71.
Vassallo MF, Camargo CA. Potential mechanisms for the hypothesized link between sunshine, vitamin D, and food allergy in children. J Allergy Clin Immunol. 2010;126:217–22.
Keet CA, Matsui EC, Savage JH. Potential mechanisms for the association between fall birth and food allergy. Allergy. 2012;67:777–82.
Haussler MR, Haussler CA, Bartik L, Whitfield GK, Hsieh J-C, Slater S, Jurutka PW. Vitamin D receptor: molecular signaling and actions of nutritional ligands in disease prevention. Nutr Rev. 2008;66:S98–112.
Gynther PP, Toropainen SS, Matilainen JMJ, Seuter SS, Carlberg CC, Väisänen SS. Mechanism of 1α,25-dihydroxyvitamin D(3)-dependent repression of interleukin-12B. ACTA-BIOENERG. 2011;1813:810–8.
Mahon B, Wittke A, Weaver V, Cantorna M. The targets of vitamin D depend on the differentiation and activation status of CD4 positive T cells. J Cell Biochem. 2003;89:922–32.
Bailey LB, Berry RJ. Folic acid supplementation and the occurrence of congenital heart defects, orofacial clefts, multiple births, and miscarriage. Am J Clin Nutr. 2005;81:1213S–7.
Matsui EC, Matsui W. Higher serum folate levels are associated with a lower risk of atopy and wheeze. J Allergy Clin Immunol. 2009;123:1253–9. e2.
Dunstan JA, West C, McCarthy S, Metcalfe J, Meldrum S, Oddy WH, Tulic MK, D'Vaz N, Prescott SL. The relationship between maternal folate status in pregnancy, cord blood folate levels, and allergic outcomes in early childhood. Allergy. 2011. doi:10.1111/j.1398-9995.2011.02714.x.
Hollingsworth JW, Maruoka S, Boon K, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–9.
Steegers-Theunissen RP, Obermann-Borst SA, Kremer D, Lindemans J, Siebel C, Steegers EA, Slagboom PE, Heijmans BT. Periconceptional Maternal Folic Acid Use of 400 μg per Day Is Related to Increased Methylation of the IGF2 Gene in the Very Young Child. PLoS ONE. 2009;4:e7845.
Ulrich CM, Toriola AT, Koepl LM, Sandifer T, Poole EM, Duggan C, McTiernan A, Issa J-PJ. Metabolic, hormonal and immunological associations with global DNA methylation among postmenopausal women. Epigenetics. 2012;7:1–9.
Cooper WN, Khulan B, Owens S, et al. DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: results of a pilot randomized controlled trial. FASEB J. 2012;26:1782–90.
Boeke CE, Baccarelli A, Kleinman KP, Burris HH, Litonjua AA, Rifas-Shiman SL, Tarantini L, Gillman MW. Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood Prospective results from a folate-replete population. Epigenetics. 2012;7:253–60.
Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, Kaur M, Vishwanatha JK, Santella RM, Cardarelli R. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr. 2011;141:1165–71.
Perera F, Tang W-Y, Herbstman J, Tang D, Levin L, Miller R. Ho S-M Relation of DNA Methylation of 5'-CpG Island of ACSL3 to Transplacental Exposure to Airborne Polycyclic Aromatic Hydrocarbons and Childhood Asthma. PLoS ONE. 2009;4:e4488.
Liu J, Ballaney M, Al-alem U, Quan C, Jin X, Perera F, Chen L-C. Miller RL Combined Inhaled Diesel Exhaust Particles and Allergen Exposure Alter Methylation of T Helper Genes and IgE Production In Vivo. Toxicol Sci. 2008;102:76–81.
Nadeau K, McDonald-Hyman C, Noth EM, Pratt B, Hammond SK, Balmes J, Tager I. Ambient air pollution impairs regulatory T-cell function in asthma. J Allergy Clin Immunol. 2010;126:845–52. e10.
Ito K, Caramori G, Lim S, Oates T, Chung KF, Barnes PJ, Adcock IM. Expression and activity of histone deacetylases in human asthmatic airways. Am J Respir Crit Care Med. 2002;166:392–6.
Cosio BG, Mann B, Ito K, Jazrawi E, Barnes PJ, Chung KF, Adcock IM. Histone acetylase and deacetylase activity in alveolar macrophages and blood mononocytes in asthma. Am J Resp Crit Care. 2004;170:141–7.
• Liang Y, Wang P, Zhao M, Liang G, Yin H, Zhang G, Wen H, Lu Q. Demethylation of the FCER1G promoter leads to FcεRI overexpression on monocytes of patients with atopic dermatitis. Allergy. 2012;67:424–30. Of importance. A key methylation event associated with the IgE pathway is described by the authors. They use a luciferase reporter assay to demonstrate the functional effects on gene expression.
Han J, Park S-G, Bae J-B, Choi J, Lyu J-M, Park SH, Kim HS, Kim Y-J, Kim S, Kim T-Y. The characteristics of genome-wide DNA methylation in naive CD4+ T cells of patients with psoriasis or atopic dermatitis. Biochem Bioph Res Co. 2012;422:157–63.
Isidoro-Garcia M, Davila-Gonzalez I. Interactions between genes and the environment. Epigenetics in allergy. Allergol Immunopathol (Madr). 2007;35:254–8.
Toperoff G, Aran D, Kark JD, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–83.
Disclosure
Dr. Prescott has received grant support from the National Health and Medical Research Council and Ilhan Foundation and served on boards for Nestle, Danone, and ALK-Abello.
Dr. Martino reported no potential conflicts of interest relevant to this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martino, D.J., Prescott, S.L. Progress in Understanding the Epigenetic Basis for Immune Development, Immune Function, and the Rising Incidence of Allergic Disease. Curr Allergy Asthma Rep 13, 85–92 (2013). https://doi.org/10.1007/s11882-012-0312-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11882-012-0312-1