Abstract
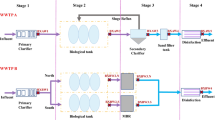
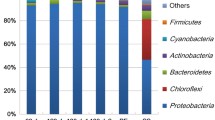
This study employed 454-pyrosequencing to investigate microbial and pathogenic communities in two wastewater reclamation and distribution systems. A total of 11972 effective 16S rRNA sequences were acquired from these two reclamation systems, and then designated to relevant taxonomic ranks by using RDP classifier. The Chao index and Shannon diversity index showed that the diversities of microbial communities decreased along wastewater reclamation processes. Proteobacteria was the most dominant phylum in reclaimed water after disinfection, which accounted for 83% and 88% in two systems, respectively. Human opportunistic pathogens, including Clostridium, Escherichia, Shigella, Pseudomonas and Mycobacterium, were selected and enriched by disinfection processes. The total chlorine and nutrients (TOC, NH3-N and NO3-N) significantly affected the microbial and pathogenic communities during reclaimed water storage and distribution processes. Our results indicated that the disinfectant-resistant pathogens should be controlled in reclaimed water, since the increases in relative abundances of pathogenic bacteria after disinfection implicate the potential public health associated with reclaimed water.
Similar content being viewed by others
References
McCarty P L, Bae J, Kim J. Domestic wastewater treatment as a net energy producer—Can this be achieved? Environmental Science & Technology, 2011, 45(17): 7100–7106
Zhang W L, Wang C, Li Y, Wang P F, Wang Q, Wang D W. Seeking sustainability: multiobjective evolutionary optimization for urban wastewater reuse in China. Environmental Science & Technology, 2014, 48(2): 1094–1102
Jemba P K, Weinrich L A, Cheng W, Giraldo E, LeChevallier M W. Regrowth of potential opportunistic pathogens and algae in reclaimed-water distribution systems. Applied and Environmental Microbiology, 2010, 76(13): 4169–4178
Cai L, Zhang T. Detecting human bacterial pathogens in wastewater treatment plants by high-throughput shotgun sequencing technique. Environmental Science & Technology, 2013, 47(10): 5433–5441
Thayanukul P, Kurisu F, Kasuga I, Furumai H. Evaluation of microbial regrowth potential by assimilable organic carbon in various reclaimed water and distribution systems. Water Research, 2013, 47(1): 225–232
Gou M, Zeng J, Wang H Z, Tang Y Q, Shigematsu T, Morimura S, Kida K. Microbial community structure and dynamics of starch-fed and glucose-fed chemostats during two years of continuous operation. Frontiers of Environmental Science & Engineering, 2015
Breitbart M, Hoare A, Nitti A, Siefert J, Haynes M, Dinsdale E, Edwards R, Souza V, Rohwer F, Hollander D. Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Ciénegas, Mexico. Environmental Microbiology, 2009, 11 (11): 16–34
DeLong E F, Preston C M, Mincer T, Rich V, Hallam S J, Frigaard N U, Martinez A, Sullivan M B, Edwards R, Brito B R, Chisholm S W, Karl D M. Community genomics among stratified microbial assemblages in the ocean’s interior. Science, 2006, 311 (5760): 496–503
Hu M, Wang X, Wen X, Xia Y. Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresource Technology, 2012, 117: 72–79
Hong P Y, Hwang C, Ling F, Andersen G L, LeChevallier M W, Liu WT. Pyrosequencing analysis of bacterial biofilm communities in water meters of a drinking water distribution system. Applied and Environmental Microbiology, 2010, 76(16): 5631–5635
Kwon S, Moon E, Kim T S, Hong S, Park H D. Pyrosequencing demonstrated complex microbial communities in a membrane filtration system for a drinking water treatment plant. Microbes and Environments, 2011, 26(2): 149–155
Pinto A J, Xi C W, Raskin L. Bacterial community structure in the drinking water microbiome is governed by filtration processes. Environmental Science & Technology, 2012, 46(16): 8851–8859
Fang Y, Xu M, Chen X, Sun G, Guo J, Wu W, Liu X. Modified pretreatment method for total microbial DNA extraction from contaminated river sediment. Frontiers of Environmental Science & Engineering, 2014, 9(3): 444–452
McKenna P, Hoffmann C, Minkah N, Aye P P, Lackner A, Liu Z, Lozupone C A, Hamady M, Knight R, Bushman F D. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathogens, 2008, 4(2): e20
Bibby K, Viau E, Peccia J. Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Research, 2010, 44(14): 4252–4260
Ye L, Zhang T. Pathogenic bacteria in sewage treatment plants as revealed by 454 pyrosequencing. Environmental Science & Technology, 2011, 45(17): 7173–7179
Liu L, You Q, Gibson V, Huang X, Chen S, Ye Z, Liu C. Treatment of swine wastewater in aerobic granular reactors: comparison of different seed granules as factors. Frontiers of Environmental Science & Engineering, 2015, 9(6): 1139–1148
Ye L, Zhang T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Applied and Environmental Microbiology, 2013, 97(6): 2681–2690
Xia S, Duan L, Song Y, Li J, Piceno Y M, Andersen G L, Alvarez-Cohen L, Moreno-Andrade I, Huang C L, Hermanowicz S W. Bacterial community structure in geographically distributed biological wastewater treatment reactors. Environmental Science & Technology, 2010, 44(19): 7391–7396
Kopilovic B, Ucakar V, Koren N, Krek M, Kraigher A. Waterborne outbreak of acute gastroenteritis in a costal area in Slovenia in June and July 2008. European Communicable Disease Bulletin, 2008, 13(34): 717–727
Moreno Y, Alonso J L, Botella S, Ferrús M A, Hernández J. Survival and injury of Arcobacter after artificial inoculation into drinking water. Research in Microbiology, 2004, 155(9): 726–730
Carlson P E Jr, Walk S T, Bourgis A E T, Liu MW, Kopliku F, Lo E, Young V B, Aronoff D M, Hanna P C. The relationship between phenotype, ribotype, and clinical disease in human Clostridium difficile isolates. Anaerobe, 2013, 24(12): 109–116
Omidbakhsh N. Evaluation of sporicidal activities of selected environmental surface disinfectants: Carrier tests with the spores of Clostridium difficile and its surrogates. American Journal of Infection Control, 2010, 38(9): 718–722
Chang J J, Chou C H, Ho C Y, Chen W E, Lay J J, Huang C C. Syntrophic co-culture of aerobic Bacillu and anaerobic Clostridium for bio-fuels and bio-hydrogen production. International Journal of Hydrogen Energy, 2008, 33(19): 5137–5146
Tortoli E. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clinical Microbiology Reviews, 2003, 16(2): 319–354
Kim C J, Kim N H, Song K H, Choe P G, Kim E S, Park S W, Kim H B, Kim N J, Kim E C, Park W B, Oh M. Differentiating rapid-and slow-growing mycobacteria by difference in time to growth detection in liquid media. Diagnostic Microbiology and Infectious Disease, 2012, 75(1): 73–76
Rastogi N, Frehel C, Ryter A, Ohayon H, Lesourd M, David H L. Multiple drug resistance in Mycobacterium avium: Is the wall architecture responsible for exclusion of antimicrobial agents? Antimicrobial Agents and Chemotherapy, 1981, 20(5): 666–677
Falkinham J OIII. The biology of environmental mycobacteria. Environmental Microbiology Reports, 2009, 1(6): 477–487
Thomas V, Loret J F, Jousset M, Greub G. Biodiversity of amoebae and amoebae-resisting bacteria in a drinking water treatment plant. Environmental Microbiology, 2008, 10(10): 2728–2745
Donnenberg M S. Shigella and enteroinvasive Escherichia coli: paradigms for pathogen evolution and host–parasite interactions. In: Escherichia Coli. 2 nd ed. Boston: Academic Press, 2013, 215–245
Huang J J, Hu H Y,Wu Y H, Wei B, Lu Y. Effect of chlorination and ultraviolet disinfection on tetA-mediated tetracycline resistance of Escherichia coli. Chemosphere, 2013, 90(8): 2247–2253
Templeton M R, Oddy F, Leung W, Rogers M. Chlorine and UV disinfection of ampicillin-resistant and trimethoprim-resistant Escherichia coli. Canadian Journal of Civil Engineering, 2009, 36(5): 889–894
Williams M M, Santo Domingo J W, Meckes M C. Population diversity in model potable water biofilms receiving chlorine or chloramine residual. Biofouling, 2005, 21(5–6): 279–288
Simões L C, Simões M, Vieira M J. Influence of the diversity of bacterial isolates from drinking water on resistance of biofilms to disinfection. Applied and Environmental Microbiology, 2010, 76 (19): 6673–6679
Lehtola M J, Miettinen I T, Vartiainen T, Myllykangas T, Martikainen P J. Microbially available organic carbon, phosphorus, and microbial growth in ozonated drinking water. Water Research, 2001, 35(7): 1635–1640
McCoy S T, VanBriesen J M. Emporal variability of bacterial diversity in a chlorinated drinking water distribution system. Journal of Environmental Engineering, 2012, 138(7): 786–795
Zeng D N, Fan Z Y, Chi L, Wang X, Qu W D, Quan Z X. Analysis of the bacterial communities associated with different drinking water treatment processes. World Journal of Microbiology & Biotechnology, 2013, 29(9): 1573–1584
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lin, Y., Li, D., Zeng, S. et al. Changes of microbial composition during wastewater reclamation and distribution systems revealed by high-throughput sequencing analyses. Front. Environ. Sci. Eng. 10, 539–547 (2016). https://doi.org/10.1007/s11783-016-0830-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11783-016-0830-5