Abstract
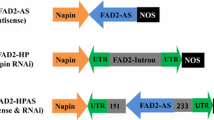
Antisense (AS) and hairpin (HP) RNA interference (RNAi) targeted gene suppression technologies have been used to modify seed oil composition. Larger numbers of AS transgenics have to be screened to achieve a targeted level of suppression compared to RNAi. We hypothesized combining AS with RNAi might result in enhanced gene suppression compared to either method individually. AS and HP-RNAi were combined as hairpin antisense (HPAS) constructs containing ~125 bp sense and antisense portions of an untranslated region of the target gene separated by an intron containing an antisense copy of a portion of the target coding region. The Δ12-desaturase FAD2, the ω3-desaturase FAD3 and β-ketoacyl-ACP synthase (KAS) II were targeted in Arabidopsis to evaluate changes in oil composition with AS, HP and HPAS constructs driven by the phaseolin promoter. Modest but statistically significant enhancements in oilseed phenotypes were observed with HPAS relative to AS and HP-RNAi. Phenotypes for HPAS suppression of FAD2 and FAD3 were indistinguishable from their strongest mutant alleles. Our data suggest that HPAS may be useful for: (1) achieving levels of suppression comparable to those of gene knockouts in a tissue specific manner. (2) Maximizing suppression of suboptimal RNAi constructs and (3) minimizing the screening of transgenics to achieve desired oilseed composition.






Similar content being viewed by others
Abbreviations
- FAD2:
-
Oleate desaturase
- FAD3:
-
Linoleate desaturase
- KASII:
-
β-Ketoacyl-ACP synthase
- Oleic acid:
-
18:1
- Stearic acid:
-
18:0
- SCD:
-
Stearoyl-CoA-desaturase
References
Napier JA (2007) The production of unusual fatty acids in transgenic plants. Annu Rev Plant Biol 58:295–319
Knutzon DS, Thompson GA, Radke SE, Johnson WB, Knauf VC, Kridl JC (1992) Modification of Brassica seed oil by antisense expression of a stearoyl-acyl carrier protein desaturase gene. Proc Natl Acad Sci USA 89:2624–2628
Damude HG, Kinney AJ (2008) Engineering oilseeds to produce nutritional fatty acids. Physiol Plant 132:1–10
Kinney AJ, Cahoon EB, Hitz WD (2002) Manipulating desaturase activities in transgenic crop plants. Biochem Soc Trans 30:1099–1103
Somerville CR, Browse JA (1991) Plant lipids: metabolism, mutants, and membranes. Science 252:80–87
Pidkowich MS, Nguyen HT, Heilmann I, Ischebeck T, Shanklin J (2007) Modulating seed beta-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc Natl Acad Sci USA 104:4742–4747
Sellwood C, Slabas AR, Rawsthorne S (2000) Effects of manipulating expression of acetyl-CoA carboxylase I in Brassica napus L. embryos. Biochem Soc Trans 28:598–600
Jadhav A, Katavic V, Marillia EF, Michael Giblin E, Barton DL, Kumar A, Sonntag C, Babic V, Keller WA, Taylor DC (2005) Increased levels of erucic acid in Brassica carinata by co-suppression and antisense repression of the endogenous FAD2 gene. Metab Eng 7:215–220
Singh S, Green A, Stoutjesdijk P, Liu Q (2000) Inverted-repeat DNA: a new gene-silencing tool for seed lipid modification. Biochem Soc Trans 28:925–927
Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408:325–330
Simons RW, Kleckner N (1988) Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet 22:567–600
Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407:319–320
Jorgensen RA (1995) Cosuppression flower color patterns, and metastable gene expression states. Science 268:686–691
Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95:13959–13964
Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952
Mol JN, van der Krol AR, van Tunen AJ, van Blokland R, de Lange P, Stuitje AR (1990) Regulation of plant gene expression by antisense RNA. FEBS Lett 268:427–430
Corey DR (2007) RNA learns from antisense. Nat Chem Biol 3:8–11
Dean NM, Bennett CF (2003) Antisense oligonucleotide-based therapeutics for cancer. Oncogene 22:9087–9096
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Stuitje AR, Verbree EC, van der Linden KH, Mietkiewska EM, Nap JP, Kneppers TJ (2003) Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol J 1:301–309
Slightom JL, Sun SM, Hall TC (1983) Complete nucleotide sequence of a French bean storage protein gene: Phaseolin. Proc Natl Acad Sci USA 80:1897–1901
van der Geest AH, Hall TC (1997) The beta-phaseolin 5′ matrix attachment region acts as an enhancer facilitator. Plant Mol Biol 33:553–557
Butte W, Eilers J, Hirsch K (1982) Trialkylsulfonium-hydroxides and trialkylselonium-hydroxides for the pyrolytic alkylation of acidic compounds. Anal Lett 15:841–850
Yamamoto K, Shibahara A, Nakayama T, Kajimoto G (1991) Determination of double-bond positions in methylene-interrupted dienoic fatty acids by GC-MS as their dimethyl disulfide adducts. Chem Phys Lipids 60:39–50
Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6:147–158
Buhr T, Sato S, Ebrahim F, Xing A, Zhou Y, Mathiesen M, Schweiger B, Kinney A, Staswick P (2002) Ribozyme termination of RNA transcripts down-regulate seed fatty acid genes in transgenic soybean. Plant J 30:155–163
Arondel V, Lemieux B, Hwang I, Gibson S, Goodman HM, Somerville CR (1992) Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 258:1353–1355
Shah S, Xin Z, Browse J (1997) Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol 114:1533–1539
Miquel M, James D Jr, Dooner H, Browse J (1993) Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc Natl Acad Sci USA 90:6208–6212
McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development not photosynthesis in an Arabidopsis mutant. Plant Cell 8:403–416
Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG (2002) hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol 129:1723–1731
James DW, Dooner HK (1990) Isolation of EMS-induced mutants in Arabidopsis altered in seed fatty acid composition. Theor Appl Genet 80:241–245
Millar AA, Kunst L (1997) Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J 12:121–131
Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6:24–35
Dormann P, Voelker TA, Ohlrogge JB (2000) Accumulation of palmitate in Arabidopsis mediated by the acyl-acyl carrier protein thioesterase FATB1. Plant Physiol 123:637–644
Bonaventure G, Salas JJ, Pollard MR, Ohlrogge JB (2003) Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 15:1020–1033
Comai L, Henikoff S (2006) TILLING: practical single-nucleotide mutation discovery. Plant J 45:684–694
Acknowledgments
We acknowledge the Office of Basic Energy Sciences of the US Department of Energy and Dow Agrosciences for their generous support.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Nguyen, T., Shanklin, J. Altering Arabidopsis Oilseed Composition by a Combined Antisense-Hairpin RNAi Gene Suppression Approach. J Am Oil Chem Soc 86, 41–49 (2009). https://doi.org/10.1007/s11746-008-1322-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11746-008-1322-y