Abstract
Two novel Aspergillus species, one belonging to the section Terrei and the other to section Flavipedes, were isolated from hypersaline soils of The National Park of Lake Urmia (Iran) and are here described as Aspergillus iranicus and Aspergillus urmiensis. A polyphasic taxonomic approach comprising extrolite profiles, phenotypic characters and molecular data (beta-tubulin, calmodulin and ribosomal polymerase II second largest subunit gene sequences) was applied to determine their novel taxonomic status. Aspergillus iranicus (CBS 139561T) is phylogenetically related to A. carneus, A. niveus, A. allahabadii and A. neoindicus, and it can be differentiated from those species by a unique extrolite pattern (citrinin, gregatins, and a terrequinone) and its conidial colour. Aspergillus urmiensis (CBS 139558T) shares a most recent common ancestor with A. templicola. The former species produces globose vesicles, and those of A. templicola are predominantly elongate. The Aspergillus urmiensis isolates produce several uncharacterized extrolites. Two other strains obtained during this study reside in a clade, together with the type strain of A. movilensis (CCF 4410T), and are identified accordingly. Based on the phylogenetic data presented in this study, A. frequens is reduced to synonymy with A. micronesiensis and A. mangaliensis is considered to be a synonym of A. templicola.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Aspergillus was described almost 300 years ago in 1729 by Micheli (Ainsworth 1976; Pitt and Hocking 1997). Since the description of the genus, it became one of the best-known and most studied fungi. Aspergillus species are important microorganisms and can have positive and negative impacts on man. They are used in food fermentations (e.g. A. oryzae, A. sojae, A. luchuensis) and for the production of drugs and enzymes (e.g. A. terreus, A. niger). Their negative impacts include degradation of agricultural products and spoilage of food and feed, production of mycotoxins and infection of animals and humans (Klich 2002a, b; Krijgsheld et al. 2013; Gregory and Thomas 1997; Suhail et al. 2007).
Species of Aspergillus have a ubiquitous distribution and occur on decaying vegetation, soil and dust worldwide (Dyer and O’Gorman 2012). They are found in terrestrial habitats and are commonly isolated from soil (Carroll and Wicklow 1992). The cosmopolitan distribution of Aspergillus in diverse ranges of ecological niches is mainly attributed to their neutral reaction to abiotic growth conditions as they are not very selective in this respect (Krijgsheld et al. 2013). Studies on the occurrence of fungi in salterns have indicated that Aspergillus and Penicillium species are among the predominant genera in these environments (Cantrell et al. 2011). The ability to tolerate high salt concentrations is a characteristic recognized for many species of Aspergillus (Tresner and Hayes 1971).
Changes in the International Code of Nomenclature for algae, fungi and plants have led to discussions whether to split Aspergillus into multiple genera or to keep it as one genus (Samson et al. 2014; Pitt and Taylor 2014). If the proposal of Samson is followed, then the genus comprises more than 340 accepted species. Based on a combination of multilocus sequence data and morphological traits, four subgenera and 23 sections are recognized in Aspergillus (Houbraken et al. 2014; Jurjević et al. 2015). During the survey of Aspergillus species in soil, several isolates belonging to the sections Flavipedes and Terrei were obtained. These sections are phylogenetically related and belong to subgenus Circumdati (Houbraken and Samson 2011; Jurjević et al. 2015). The taxonomy of these sections has been studied in detail; however, there is confusion in section Flavipedes because two studies describing new, similar species were published online around the same time (Hubka et al. 2015; Visagie et al. 2014).
Lake Urmia, located in the northwest of Iran between the provinces East and West Azerbaijan, is the largest lake in the Middle East and the second saltiest lake in the world after the Dead Sea. The National Park of Urmia Lake is a protected area and comprises two ecosystems (water and land). The salinity of the lake ranges between 120 g/L and more than 300 g/L and the lake is surrounded by marsh lands (Asem et al. 2014). Its land ecosystem consists of 102 islands, covering an area of 7816 ha (Asem et al. 2014). During the investigation of the biodiversity of Aspergillus species inhabiting hypersaline soils of this National Park, we discovered strains belonging to the sections Terrei and Flavipedes, which did not fit into any of the described species of Aspergillus. We used a polyphasic taxonomic approach to fully characterize these novel species. The macro- and micromorphology of the isolates were examined and extrolite patterns determined. For phylogenetic analysis, partial sequences of the β-tubulin (BenA), calmodulin (CaM) and ribosomal polymerase II second largest subunit (RPB2) genes were analyzed.
Materials and methods
Isolates
Soil samples were collected at 10–15 cm depth from two islands (Aspear and Kabodan) and the coastal areas of Lake Urmia, Iran, during 2011 and 2012. Isolations were carried out using the soil dilution plate and Warcup soil plate method (Warcup 1950) on malt extract agar (MEA), glucose peptone yeast extract agar (GPY) and potato dextrose agar (PDA) culture media containing NaCl concentrations varying from 0 to 30 %. Single spore isolations were made to obtain pure cultures. Dried cultures of the types are preserved at the fungarium of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands. The living strains (Table 1) were deposited in the Culture Collection of Tabriz University (CCTU), CBS-KNAW and the internal culture collection of the Applied and Industrial Mycology group (DTO) of CBS-KNAW.
Morphological analysis
For macro-morphological observations, isolates were cultivated on Czapek yeast autolysate agar (CYA), Czapek agar (CZA), yeast extract sucrose agar (YES), oatmeal agar (OA) (medium compositions according to Samson and Frisvad 2004) and malt extract agar (2 % MEA; Merck, Germany). The isolates were inoculated at three points on each plate of each medium and incubated at 25 °C and 37 °C in the dark. Colony growth characteristics were recorded after 7 days of incubation. Colour names and numeric codes used in the description refer to Klich (2002b). For microscopic observations, mounts were made in lactic acid from colonies grown on MEA; a drop of alcohol was added to remove air bubbles and excess conidia. For micro-morphological examination, light microscopy (Olympus BH2) was employed. Photographs were captured using a Olympus-BX41 light microscope with an Olympus digital camera system (DP 25).
Extrolite analysis
Cultures were grown for 7 days on CYA and YES agar prior extrolite extraction. Three agar plugs were extracted per agar medium as described before (Houbraken et al. 2012; Nielsen et al. 2011). The extracts were analysed using UHPLC-DAD and compounds were identified against an internal database of UV spectra and literature. Standards were available for the extrolites reported in Nielsen et al. (2011).
Phylogenetic analyses
Strains were grown for 3–10 days on MEA prior to DNA extraction. Genomic DNA was extracted using the Ultraclean™ Microbial DNA isolation kit (MoBio, Solana Beach, USA). After DNA extraction, parts of the BenA, CaM, internal transcribed spacers (ITS) and RPB2 regions were amplified, sequenced and annotated (Houbraken et al. 2012; Houbraken and Samson 2011). The newly generated sequences were supplemented with sequence data from GenBank. After compilation of the sequence data sets, all datasets were aligned using the MAFFT multiple sequence alignment software v.7.221 (Katoh and Standley 2013). The best model for the maximum likelihood analysis was selected based on the Akaike Information Criterion (AIC), which was calculated in MEGA6. All positions containing gaps and missing data were eliminated. A ML analysis was performed on the individual and combined datasets. The individual datasets were analysed in MEGA v.6.0.6 (Tamura et al. 2013) and the combined multilocus alignment in RAxML (randomised accelerated maximum likelihood, v.7.0.4) software (Stamatakis et al. 2008). In the RAxML analysis, each dataset was treated as a separate partition. The statistical support was evaluated by 1000 bootstrap replicates. A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v.3.1.1 (Ronquist and Huelsenbeck 2003). Models of nucleotide substitution for each gene were included for each partition. The Bayesian analysis was performed with two sets of four chains (one cold and three heated) and the stoprule option, stopping the analysis at an average standard deviation of split frequencies of 0.01. Aspergillus candidus NRRL 303T (sect. Candidi) was used as an outgroup. Newly obtained sequences were deposited in GenBank, see Table 1.
Results
DNA phylogeny and identification
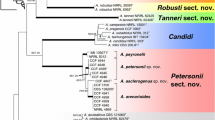
A total of 48 isolates belonging to sections Flavipedes, Terrei and Jani including the outgroup (A. candidus NRRL 303T) were included in the multigene analysis (gene boundaries of BenA: 1–554; CaM: 555–1160; ITS: 1161–1706; RPB2: 1707–2679). 2679 characters including gaps were processed, 1198 distinct alignment patterns were present and the proportion of gaps in the alignment was 7.28 %. For Bayesian analysis, a HKY+G+I model was selected for BenA, and a GTR+G+I model for the CaM and RPB2 dataset. The posterior probability values correlated well with the bootstrap supports from the ML analysis (Fig. 1).
Best-scoring Maximum Likelihood trees based on BenA, CaM, RPB2 and a combined dataset of sequences showing the relationship of species belonging to Aspergillus sections Flavipedes, Jani and Terrei. The strains in bold were isolated in this study. The bootstrap percentages of the Maximum Likelihood (ML) analysis are presented at the nodes together with the posterior probability (pp) values (ML/pp). Bootstrap values below 70 % and less than 0.95 pp are omitted or indicated as a hyphen, whereas asterisks indicate full support (100 % bootstrap or 1.00 pp). The branches with more than 95 % bootstrap support and 1.00 in the Bayesian analysis are thickened
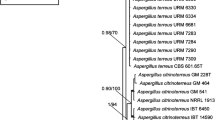
The results of the combined analysis is shown in Fig. 1 and demonstrates that the isolates can be divided into three well-supported groups, representing three sections in Aspergillus: Flavipedes, Terrei and Jani. Twelve species are currently accepted in section Flavipedes, including the novel species A. urmiensis. The type strains of A. templicola (CBS 138181T) and A. mangaliensis (CCF 4698T) form a well-supported clade, together with two other strains (CBS 138180 and CCF 869). Similarly, A. micronesiensis (CBS 138183T) and A. frequens (NRRL 4578T) fall into the same clade. Five isolates obtained in our study belong to section Flavipedes. Isolates CBS 139558, CBS 139766 and CBS 139557 form a well-supported clade in all analyses. This group of isolates is described here as Aspergillus urmiensis. Based on the combined analysis, this new species is a sister species of A. templicola. Two other isolates from Iranian soil (CBS 139559 and CBS 139562) reside in a clade together with the type strain of A. movilensis (CCF 4410T) and are identified accordingly.
The isolates CBS 139560 and CBS 139561T, described as a new species A. iranicus in this study, have identical BenA, CaM, ITS and RPB2 sequences. These isolates cluster together in all analyses and never with any of the other accepted species in section Terrei. The exact phylogenetic position of these isolates is unresolved in the CaM and RPB2 analysis, but the BenA and combined analyses show that these strains are basal to A. carneus, A. niveus, A. allahabadii and A. neoindicus.
Extrolites analysis
The extrolites profiles of the strains isolated during this study were determined. Both A. iranicus strains produced citrinin, gregatins, and a terrequinone and CBS 139560 produced an additional compound tentatively identified as asperamide. The A. urmiensis isolates had similar extrolite profiles; however, none of the detected compounds could be identified and remain uncharacterized. The two A. movilensis strains isolated in this study produced asperphenamate, aspochalasins, a butyrolactone and other unique extrolites. The phylogenetically closely related strain NRRL 4610 (= IBT 30185), which was identified as A. movilensis by Hubka et al. (2015), produced asperphenamate, a butyrolactone and a cyclic peptide resembling psychrophilin. This extrolite profile is similar to those of the two A. movilensis strains from this study.
Taxonomy
Aspergillus iranicus Arzanlou, Houbraken & Samadi, sp. nov. Mycobank MB817473. Figure 2.
Etymology: in reference to the ex-type strain, which was isolated from hypersaline soil in Iran.
Diagnosis: Phylogenetically basal to A. carneus, A. niveus, A. allahabadii and A. neoindicus. Good growth on CYA at 37 °C (34–38 mm), radiate conidial heads, accessory conidia produced. Unique extrolite profile: citrinin, gregatins, terrequinone X (maybe terrequinone A).
Typus: Iran, Urmia, Aspear Island, soil, 2012, isolated by U. Ghosta and R. Samad (holotype CBS H-22338, culture ex-type CCTU 756 = CBS 139561 = IBT 32596 = DTO 203-D7).
Additional material examined: Iran, Jade Darya (seaside), Urmia, soil, 2011, isolated by U. Ghosta and R. Samad, CCTU 750 = CBS 139560 = IBT 32595 = DTO 203-D1.
ITS barcode: KP987077 (alternative markers: BenA = KP987045; CaM = KP987060; RPB2 = KP987034).
Colony diameter (mm): 7 days, 25 °C, CYA 28–32; CZ 24–28; MEA 30–34; YES 23–27; 7 days, 37 °C, CYA37 °C 34–38; CZ37 °C 37–39; MEA37 °C 36–40; YES37 °C 36–40.
Colony characters: CYA 25 °C, 7 days: mycelium white; sclerotia absent; sporulation dense; conidial mass white, colour of the colony changed to peach (4) after 3 weeks; soluble pigment absent; colonies felt, centrally velutinous, sulcate; reverse honey (64) and sulfur yellow (15) in the deeper parts of the sulcations. YES 25 °C, 7 days: mycelium white; sclerotia absent; sporulation moderate; conidial mass white; soluble pigment absent; exudate sparse, amber (47); colony texture velvety, floccose in centre; sulcate; reverse pale luteous (11) to luteous (12) (Fig. 2). CZ 25 7 days: mycelium white at margin to greenish yellow in the centre (16); sclerotia absent; sporulation moderate in centre, conidial mass white; colonies felt, centrally floccose, sulcate; soluble pigment absent; greenish yellow (16) exudate produced after 14 days; reverse citrine (13). MEA 25 °C, 7 days: mycelium white; sclerotia absent; sporulation dense; conidial mass white; soluble pigment absent; exudate absent; colonies velutinous to lightly floccose; sulcate; reverse pale luteous (11) (Fig. 2).
Micromorphology: Stipes (375–) 550–625 (−800) × (2.5–) 4–5 (−7) μm, smooth, aseptate to occasionally septate, walls pale yellow pigmented, thick-walled (1 μm). Foot cell in two forms: symmetric and asymmetric. Conidial heads radiate on MEA, YES, CZ and radiate to loosely columnar on CYA. Vesicles (14.5–) 20–23 (−32) × (7–) 11–13 (−16) μm, spathulate, wall thickness less than 1 μm, uncoloured. Conidiophores biseriate; the fertile part covering 1/3 to 1/4 of the upper part of the vesicle, occasionally small conidiophores with diminutive heads present. Metulae (5–) 6–7 (−8) × (2–) 3 (−4) μm, cylindrical, walls smooth, uncolorued. Phialides, 1–3 on each metula, (5–) 6–7 (−9) × 2–3 μm, cylindrical tapering to a distinct collulum. Conidia 2–2.5 × 1.8–2.5 μm in diameter, globose to subglobose, smooth-walled, hyaline (Fig. 2). Accessory conidia abundant, sessile or on the short, hyaline, micronematous conidiophores bearing conidia, globose, subglobose, elliptical, clavate, commonly truncate (4–) 5–6 (−7) μm (Fig. 2).
Notes: Aspergillus iranicus is phylogenetically related to A. carneus, A. niveus, A. allahabadii and A. neoindicus; however, it can be differentiated from these species by a combination of cultural and micro-morphological characteristics. Aspergillus neoindicus produces yellow-green mycelial tufts and the mycelium of A. iranicus is white. Furthermore, the conidial colour en masse of A. iranicus is in shades of yellow and this feature is not shared with A. niveus (white) and A. carneus (vinaceous fawn). Aspergillus iranicus produces accessory conidia and those were also reported in A. terreus, A. carneus, A. niveus and A. alabamensis.
Aspergillus urmiensis Arzanlou, Houbraken & Samadi, sp. nov. Mycobank MB817474. Figure 3.
Etymology: in reference to the ex-type strain, which was isolated from soil in Urmia, West Azerbaijan province, Iran.
Diagnosis: Conidial colour on CYA, MEA and YES ochreous, good growth on CYA incubated at 37 °C (16–20 mm), vesicles subglobose to globose measuring (17–) 20–23 (−30) × (16–) 19–22 (−30) μm.
Typus: Iran, Urmia, Jade Darya (seaside), soil, 2011, isolated by U. Ghosta and R. Samadi (holotype CBS H-22671, culture ex-type CCTU 742 = CBS 139558 = IBT 32593 = DTO 203-C2).
Additional material examined: Iran, soil, 2011, isolated by U. Ghosta and R. Samad: CCTU 734 = CBS 139557 = DTO 203-B3; CCTU 743 = CBS 139766 = IBT 32598 = DTO 203-C3.
ITS barcode: KP987073 (alternative markers: BenA = KP987041; CaM = KP987056; RPB2 = KP987030).
Colony diameter (mm): 7 days, 25 °C, CYA 28–32; CZ 20–24; MEA 23–27; YES 21–24; 7 days, 37 °C, CYA37 °C 21–23; CZ37 °C 16–20; MEA37 °C 17–19; YES37 °C 18–20.
Colony characters: CYA 25 °C, 7 days: mycelium white; sporulation strong; conidial mass ochreous (44) sclerotia absent; soluble pigment luteous (7); exudate after 21 days produced; umber-coloured (9); colony texture floccose in centre to felt in margin; sulcate with low umbonate in centre; reverse sienna (8) and one umber (9) line present in middle of colony. YES 25 °C, 7 days: submerged mycelium at the margin of colony ochreous (44); white aerial mycelium appeared after 28 days; sporulation strong; conidial mass ochreous (44); sclerotia absent; soluble pigment luteous (12); exudate absent; colony texture lanose in centre to felt in margin; sulcate with umbonate in centre; reverse luteous (12) in centre pale luteous (11) in margin of colony. CZ 25 °C, 7d: mycelium white; sporulation strong, conidial mass ochreous (44); sclerotia absent; colony texture lanose; sulcate with lightly umbonate in centre; soluble pigment slightly produced, luteous; exudate absent; reverse orange (7) (Fig. 3). MEA 25 °C, 7 days: mycelium ochreous (44); sporulation strong; conidial mass ochreous (44); sclerotia absent; orange (7) uncoloured exudate after 14 days frequently produced; colony texture lanose; sulcate; reverse luteous (12) (Fig. 3).
Micromorphology: Stipes mostly hyaline close to the vesicle and light brown to brown closer to the base, (350–) 700–850 (−1330) × (5–) 8–10 (−12) μm, smooth-walled, wall thickening 1 μm, aseptate, rarely with one septum, foot cell symmetric or asymmetric, amber (47). Conidial heads radiate; vesicles (17–) 20–23 (−30) × (16–) 19–22 (−30) μm, subglobose to globose, wall thickness less than 0.8 μm, uncoloured. Conidiophores biseriate or uniseriate; Metulae (4–) 5–6 (−7.5) × (1.5–) 2–3 (−4) μm, wedge-shaped, walls smooth, uncoloured, covering 4/5 of the upper part of the vesicle. Phialides 2–5 on each metula, (2–) 5–7 (−8) × (1–) 1.5–2 (−3) μm, cylindrical, with distinct collulum. Conidia 2–3 μm, globose, smooth-walled, hyaline (Fig. 3). Accessory conidia present in relatively small numbers, sessile or on the short, hyaline, micronematous conidiophores bearing conidia, globose, subglobose, clavate, commonly truncate (4–) 5–6 (−7) μm (Fig. 3). No ascomata, ascospores or Hülle cells observed.
Notes: Aspergillus urmiensis is phylogenetically most closely related to A. templicola. The former species produces globose vesicles, and those of A. templicola are predominantly elongate. Aspergillus urmiensis can be differentiated from A. luppii, A. movilensis, A. polyporicola and A. spelaeus by larger colony diameters (16–20 vs. 0–17 mm) on CYA incubated at 37 °C. This new species can be differentiated from A. ardalensis based on the diameter of the vesicles (A. ardalensis, 18.5 μm; A. urmiensis, 22 μm). A. neoflavipes produces bright yellow colonies on CYA and MEA, and has a sexual state; both features are not observed in A. urmiensis. A. micronesiensis and A. iizukae generally produce Hülle cells and these structures were not detected in A. urmiensis (Hubka et al. 2015; Visagie et al. 2014).
Discussion
The National Park of Urmia has a unique ecosystem which consists of a range of regular to extreme environmental conditions (Asem et al. 2014). During a survey on the biodiversity of Aspergillus species inhabiting hypersaline soils of the Urmia Lake basin, we discovered strains belonging to the sections Terrei and Flavipedes, and some that could not be reliably identified to any described Aspergillus species. Species in section Terrei and Flavipedes are phenotypically related and the taxonomy of these sections based on morphology is troublesome. In the past, species currently classified in section Terrei were placed in the section Flavipedes due to overlaps in cultural and morphological characteristics (Raper and Fennell 1965; Samson 1979; Hubka et al. 2015). DNA sequencing and phylogenetic analysis has provided a reproducible and robust tool for species classification and identification in fungi including Aspergilli (Hong et al. 2005; Peterson 2008; Schoch et al. 2012; Samson et al. 2014). Sequence data from different genomic regions (eg. BenA, CaM, ITS, large ribosomal subunit (LSU) and RPB2) have been employed to delineate sections and species boundaries in Aspergillus (e.g. Peterson 2000, 2008; Varga et al. 2005; Hubka et al. 2015). The taxonomy of section Flavipedes was recently revised and 10 species were accepted (seven described as new) (A. ardalensis, A. flavipedes, A. frequens, A. iizukae, A. luppii, A. mangaliensis, A. movilensis, A. neoflavipedes, A. polyporicola and A. spelaeus) (Hubka et al. 2015). Concurrently, another three additions to this section were made (A. templicola, A. capensis and A. micronesiensis) (Visagie et al. 2014). The type strains of A. frequens (NRRL 4578T) and A. micronesiensis (CBS 138183T) shared identical BenA, CaM and RPB2 sequences. Based on these data, A. frequens (Hubka et al. 2015) is reduced here to synonymy with A. micronesiensis (Visagie et al. 2014). Furthermore, the invalidly described species Aspergillus sunderbanii (Arts 40.3, 40.4, 40.5) is also a synonym of A. micronesiensis. The type strain of A. mangaliensis CCF 4698T is phylogenetically close, but not identical, to the type of A. templicola (CBS 138181T) (similarity BenA 98.8 %; CaM 98.7 %; RPB2 99.1 %). Based on gene concordance, they could represent two separate species; however, due to the high similarity in the investigated gene regions, we treat both as conspecific. Analysing additional strains in future will generate more insight on the status of these species.
In this study, different isolates from section Flavipedes were isolated from soils with different amounts of salinity up to 70 dS/m of the seaside and islands of the National Park of Lake Urmia. Two isolates (CBS 139559 and CBS 13562) reside in a clade together with the type strain of A. movilensis (CCF 4410T) and are accordingly identified as such. The isolates CBS 139558, CBS 139766 and CBS 139557 formed a well-supported clade in both single-gene and combined phylogenetic analyses. This group of isolates is described here as a new species named Aspergillus urmiensis. A. urmiensis is phylogenetically closely related to A. templicola and can be differentiated from other species belonging to section Flavipedes by a combination of cultural and morphological characters such as growth rate on CYA incubated at 37 °C, conidial colour, the shape and diameter of the vesicles and the presence or absence of Hülle cells and/or ascomata. This species also produces accessory conidia, a feature shared with all other members of section Flavipedes (Hubka et al. 2015).
Two strains (CBS 139560 and CBS 139561) isolated during this study belong to section Terrei and form a lineage distinct from the other accepted species. Currently, 15 species are accepted in this section: A. alabamensis, A. allahabadii, A. ambiguous, A. aureoterreus, A. carneus, A. citrinoterreus, A. floccosus, A. hortai, A. microcysticus, A. neoafricanus, A. neoindicus, A. neoniveus, A. niveus, A. pseudoterreus and A. terreus (Peterson 2008; Balajee et al. 2009; Samson et al. 2011; Guinea et al. 2015). We describe the new species A. iranicus in this section based on the concordance between the BenA, CaM and RPB2 genes and the unique phylogenetic position of the isolates in section Terrei in the combined analyses. Both isolates have identical BenA, CaM, ITS and RPB2 sequences. Analysis of the CaM and RPB2 data sets could not resolve the exact phylogenetic position of these isolates, but the BenA and combined analysis show that the strains are basal to A. carneus, A. niveus, A. allahabadii and A. neoindicus. Besides the unique phylogenetic position, the A. iranicus isolates can also be differentiated from A. carneus, A. niveus, A. allahabadii and A. neoindicus by a combination of cultural and micro-morphological characteristics (see “Taxonomy”).
Members of the section Flavipedes are known from different types of soil, especially in subtropical and tropical soils. Many species in this section are adapted to reduced water activity conditions and are able to grow in natural dry habitats. For example, A. flavipes isolates tolerate relatively high concentrations of osmotically active solutes in media, being able to grow on media with 40 % (w/v) sucrose and 25 % (w/v) NaCl (Tresner and Hayes 1971; Moustafa and AL-Musallam 1975) and were isolated from natural habitats with high NaCl concentration such as salterns (Moustafa 1975; Butinar et al. 2011; Cantrell et al. 2011), brackish water (Pawar and Thirumalachar 1966) or coastal sand of the Dead Sea (Grishkhan et al. 2003). The most well-known species from section Terrei is A. terreus, a cosmopolitan species known from desert and grassland soils, compost heaps, and also as contaminants on stored corn, barley and peanuts (Kozakiewicz 1989). This and other species such as A. alabamensis, A. citrinoterreus and A. hortai are also clinically significant (Balajee et al. 2009). In the present study, A. iranicus is described as new species in this section from hypersaline soils of the Urmia Lake basin. There is no report available on the tolerance of species in section Terrei to high concentrations of osmotically active solutes in media.
Extrolite profile analyses revealed that A. iranicus isolates produce citrinin, gregatins, and a terrequinone in common, and isolate CBS 139560 additionally produces an extrolite tentatively identified as asperamide. The hepatotoxic extrolite citrinin is also known from several other species in this section, namely, A. alabamensis, A. allahabadii, A. carneus, A. floccosus, A. hortai, A. neoindicus, A. niveus and A. pseudoterreus (Samson et al. 2011). A diverse array of metabolites, including acetylaranotin, asperphenamate, aspochalamins, aspulvinones, asteltoxin, asterric acid, asterriquinones, aszonalenins, atrovenetins, butyrolactones, citreoisocoumarins, citreoviridins, citrinins, decaturins, fulvic acid, geodins, gregatins, mevinolins, serantrypinone, terreic acid (only the precursor 3,6-dihydroxytoluquinone found), terreins, terrequinones, terretonins and territrems, are known from section Terrei species (Samson et al. 2011). Two additional metabolites namely gregatins and a compound tentatively identified as asperamide were found in A. iranicus and these compounds are new for the section Terrei. Members of the section Flavipedes are rich producers of bioactive secondary metabolites, some of which possess biotechnological and pharmacological significance (Hubka et al. 2015). Aspergillus flavipes is well studied with respect to extrolite production. A wide array of bioactive compounds is reported to be produced by this species and was listed by Hubka et al. (2015). Aspergillus movilensis CBS 139559 produces asperphenamate, aspochalasins, a butyrolactone and other unique extrolites. Strain NRRL 4610 (=IBT 30185) identified as A. movilensis by Hubka et al. (2015) produces asperphenamate, a butyrolactone and a cyclic peptide resembling psychrophilin, and this pattern of extrolites is very similar to that of CBS 139559. The A. urmiensis isolates (CBS 139558, CBS 139766 and CBS 139557) have similar extrolite profiles and produce several uncharacterised extrolites. These extrolites did not match with any of the known secondary metabolites in Aspergillus and might represent novel bioactive compounds. These compounds need structure elucidation and can be further evaluated on their pharmacological and biotechnological significance.
References
Ainsworth GC (1976) Introduction to the history of mycology. Cambridge University Press, Cambridge
Asem A., Eimanifar A., Djamal M., De los Rios P. and WinkM. (2014) Biodiversity of the Hypersaline Urmia Lake National Park (NW Iran). Diversity 6:102–132
Balajee SA, Baddley JW, Peterson SW, Nickle D, Varga J, Boey A, Lass-Flōrl C, Frisvad JC, Samson RA (2009) Aspergillus alabamensis, a new clinically relevant species in the section Terrei. Eukaryot Cell 8:713–722
Butinar L, Frisvad JC, Gunde-Cimerman N (2011) Hypersaline waters a potential source of foodborne toxigenic aspergilli and penicillia. FEMS Microbiol Ecol 77:186–199
Cantrell SA, Dianese JC, Fell J, Gunde-Cimerman N, Zalar P (2011) Unusual fungal niches. Mycologia 103:1161–1174
Carroll GC, Wicklow DT (eds) (1992) The fungal community, its organization and role in the ecosystem, 2nd edn. Marcel Dekker, New York
Dyer PS, O’Gorman CM (2012) Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev 36:165–192
Gregory S, Thomas HA (1997) The importance of fungi to man. Genome Res 7:1041–1044
Grishkhan I, Nevo E, Wasser SP (2003) Soil micromycete diversity in the hypersaline Dead Sea coastal area, Israel. Mycol Prog 2:19–28
Guinea J, Sandoval-Denis M, Escribano P, Peláez T, Guarro J, Bouza E (2015) Aspergillus citrinoterreus, a new species of section Terrei isolated from samples of patients with nonhematological predisposing conditions. J Clin Microbiol 53:611–617
Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA (2005) Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 97:1316–1329
Houbraken J, Samson RA (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol 70:1–51
Houbraken J, Frisvad JC, Seifert KA, Overy DP, Tuthill DM, Valdez JG, Samson RA (2012) New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia 29:78–100
Houbraken J, de Vries RP, Samson RA (2014) Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol 86:199–249
Hubka V, Kolarik M, Novakova A, Jurjević Ž (2015) Revision of Aspergillus section Flavipedes: Seven new species and proposal of section Jani sect. nov. Mycologia 107:169–208
Jurjević Ž, Kubátová A, Kolarik M, Hubka V (2015) Taxonomy of Aspergillus section Petersonii sect. nov. encompassing indoor and soil-borne species with predominant tropical distribution. Plant Syst Evol 301:2441–2462
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780
Klich MA (2002a) Biogeography of Aspergillus species in soil and litter. Mycologia 94:21–27
Klich MA (2002b) Identification of common Aspergillus species. Centraalbureau voor Schimmelcultures. Utrecht, The Netherlands
Kozakiewicz Z (1989) Aspergillus species on stored products. Mycol Pap 161:1–188
Krijgsheld P, Bleichrodt RJ, van Veluw JG, Wang F, Müller WG, Dijksterhuis J, Wösten HAB (2013) Development in Aspergillus. Stud Mycol 74:1–29
Moustafa AF (1975) Osmophilous fungi in the salt of Kuwait. Can J Microbiol 21:1573–1580
Moustafa AF, AL-Musallam AA (1975) Contribution to the fungal flora of Kuwait. Trans Br Mycol Soc 65:547–553
Nielsen KF, Månsson M, Rank C, Frisvad JC, Larsen TO (2011) Dereplication of microbial natural products by LC-DAD-TOFMS. J Nat Prod 74:2338–2348
Pawar VH, Thirumalachar MG (1966) Studies on halophilic soil fungi from Bombay. Nova Hedwigia 12:497–508
Peterson SW (2000) Phylogenetic relationships in Aspergillus based on rDNA sequence analysis. In: Samson RA, Pitt JI (eds) Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood, Amsterdam, pp 323–355
Peterson SW (2008) Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226
Pitt JI, Hocking AD (1997) Fungi and food spoilage, 2nd edn. Blackie, London
Pitt JI, Taylor JW (2014) Aspergillus, its sexual states and the new International Code of Nomenclature. Mycologia 106:1051–1062
Raper KB, Fennell DI (1965) The genus Aspergillus. Williams & Wilkins, Baltimore
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Samson RA (1979) A compilation of the Aspergilli described since 1965. Stud Mycol 18:1–38
Samson RA, Frisvad JC (2004) Penicillium subgenus Penicillium: new taxonomic schemes, mycotoxins and other extrolites. Stud Mycol 49:1–157
Samson RA, Peterson SW, Frisvad JC, Varga J (2011) New species in Aspergillus section Terrei. Stud Mycol 69:39–55
Samson RA, Visagie CM, Houbraken J, Hong S-B, Hubka V, Klaassen CHW, Perrone G, Seifert KA, Susca A, Tanney JB, Varga J, Kocsubé S, Szigeti G, Yaguchi Y, Frisvad JC (2014) Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 78:141–178
Schoch CL, Seifert KA, Huhndorf S, Spouge RV, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109:6241–6246
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771
Suhail M, Irum M, Jatt T, Korejo F, Abro H (2007) Aspergillus mycoflora isolated from soil of kotri barrage sindh. Pak J Bot 39:981–984
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Tresner HD, Hayes JA (1971) Sodium chloride tolerance of terrestrial fungi. Appl Microbiol 22:210–213
Varga J, Tóth B, Kocsubé S, Farkas B, Szakács G, Téren J, Kozakiewicz Z (2005) Evolutionary relationships among Aspergillus terreus isolates and their relatives. Antonie Van Leeuwenhoek 88:141–150
Visagie CM, Houbraken J, Frisvad JC et al (2014) Identification and nomenclature of the genus Penicillium. Stud Mycol 78:343–371
Warcup JH (1950) The soil-plate method for isolation of fungi from soil. Nature 166:117–118
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editors: Pedro W. Crous and Roland Kirschner
This article is part of the Special Issue Biodiversity of Hyphomycetes - Special Issue in honor of Dr. Subramanian.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Arzanlou, M., Samadi, R., Frisvad, J.C. et al. Two novel Aspergillus species from hypersaline soils of The National Park of Lake Urmia, Iran. Mycol Progress 15, 1081–1092 (2016). https://doi.org/10.1007/s11557-016-1230-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-016-1230-8