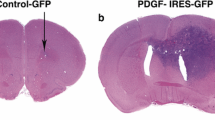
Abstract
Paracrine PDGF signaling is involved in many processes in the body, both normal and pathological, including embryonic development, angiogenesis, and wound healing as well as liver fibrosis, atherosclerosis, and cancers. We explored this seemingly dual (normal and pathological) role of PDGF mathematically by modeling the release of PDGF in brain tissue and then varying the dynamics of this release. Resulting simulations show that by varying the dynamics of a PDGF source, our model predicts three possible outcomes for PDGF-driven cellular recruitment and lesion growth: (1) localized, short duration of growth, (2) localized, chronic growth, and (3) widespread chronic growth. Further, our model predicts that the type of response is much more sensitive to the duration of PDGF exposure than the maximum level of that exposure. This suggests that extended duration of paracrine PDGF signal during otherwise normal processes could potentially lead to lesions having a phenotype consistent with pathologic conditions.



Similar content being viewed by others
References
Allamargot C, Pouplard-Barthelaix A, Fressinaud C (2001) A single intracerebral microinjection of platelet-derived growth factor (PDGF) accelerates the rate of remyelination in vivo. Brain Res 918(1):28–39. https://doi.org/10.1016/S0006-8993(01)02761-5
Andrae J, Gallini R, Betsholtz C (2008) Role of platelet-derived growth factors in physiology and medicine. Genes Dev 22(10):1276–1312. https://doi.org/10.1101/gad.1653708
Andrew J, Hoyland J, Freemont A, Marsh D (1995) Platelet-derived growth factor expression in normally healing human fractures. Bone 16(4):455–460. https://doi.org/10.1016/8756-3282(95)90191-4
Apperley JF, Gardembas M, Melo JV, Russell-Jones R, Bain BJ, Baxter EJ, Chase A, Chessells JM, Colombat M, Dearden CE, Dimitrijevic S, Mahon FX, Marin D, Nikolova Z, Olavarria E, Silberman S, Schultheis B, Cross NC, Goldman JM (2002) Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med 347(7):481–487. https://doi.org/10.1056/NEJMoa020150
Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P (2006) Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci 26(25):6781–90. https://doi.org/10.1523/jneurosci.0514-06.2006
Assanah MC, Bruce JN, Suzuki SO, Chen A, Goldman JE, Canoll P (2009) Pdgf stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia. https://doi.org/10.1002/glia.20895
Bates KA, Drummond ES, Cozens GS, Harvey AR (2016) Vascular insufficiency, not inflammation, contributes to chronic gliosis in a rat CNS transplantation model. Restor Neurol Neurosci 34(2):313–323. https://doi.org/10.3233/RNN-150591
Betsholtz C (2003) Biology of platelet-derived growth factors in development. Birth Defects Res Part C Embryo Today Rev 69(4):272–285. https://doi.org/10.1002/bdrc.10030
Bissell L, Tibrewal S, Sahni V, Khan WS (2015) Growth factors and platelet rich plasma in anterior cruciate ligament reconstruction. Curr Stem Cell Res Therapy 10(1):19–25
Blinkov SM, Glezer II (1968) The human brain in figures and tables: a quantitative handbook. Basic Books, New York
Boor P, Ostendorf T, Floege J (2014) PDGF and the progression of renal disease. Nephrol Dial Transplant 29(suppl 1):i45–i54. https://doi.org/10.1093/ndt/gft273
Boucher P, Gotthardt M (2004) LRP and PDGF signaling: a pathway to atherosclerosis. Trends Cardiovasc Med 14(2):55–60. https://doi.org/10.1016/j.tcm.2003.12.001
Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, Odell MM, Bauer RL, Ren HP, Haugen HS, Yeh MM, Fausto N (2005) Platelet-derived growth factor c induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci USA 102(9):3389–3394. https://doi.org/10.1073/pnas.0409722102
Chintalgattu V, Ai D, Langley RR, Zhang J, Bankson JA, Shih TL, Reddy AK, Coombes KR, Daher IN, Pati S, Patel SS, Pocius JS, Taffet GE, Buja LM, Entman ML, Khakoo AY (2010) Cardiomyocyte PDGFR-signaling is an essential component of the mouse cardiac response to load-induced stress. J Clin Investig 120(2):472–484. https://doi.org/10.1172/JCI39434
De Wever O, Demetter P, Mareel M, Bracke M (2008) Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer 123(10):2229–2238. https://doi.org/10.1002/ijc.23925
Demoulin JB, Essaghir A (2014) PDGF receptor signaling networks in normal and cancer cells. Cytokine Growth Factor Rev 25(3):273–283. https://doi.org/10.1016/j.cytogfr.2014.03.003
Eschenhagen T, Force T, Ewer MS, de Keulenaer GW, Suter TM, Anker SD, Avkiran M, de Azambuja E, Balligand JL, Brutsaert DL, Condorelli G, Hansen A, Heymans S, Hill JA, Hirsch E, Hilfiker-Kleiner D, Janssens S, de Jong S, Neubauer G, Pieske B, Ponikowski P, Pirmohamed M, Rauchhaus M, Sawyer D, Sugden PH, Wojta J, Zannad F, Shah AM (2011) Cardiovascular side effects of cancer therapies: a position statement from the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail 13(1):1–10. https://doi.org/10.1093/eurjhf/hfq213
Farin A, Suzuki SO, Weiker M, Goldman JE, Bruce JN, Canoll P (2006) Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia 53(8):799–808. https://doi.org/10.1002/glia.20334
Fomchenko EI, Holland EC (2007) Platelet-derived growth factor-mediated gliomagenesis and brain tumor recruitment. Neurosurg Clin N Am 18(1):39–58. https://doi.org/10.1016/j.nec.2006.10.006
Frost EE, Zhou Z, Krasnesky K, Armstrong RC (2009) Initiation of oligodendrocyte progenitor cell migration by a PDGF-A activated extracellular regulated kinase (ERK) signaling pathway. Neurochem Res 34(1):169–181. https://doi.org/10.1007/s11064-008-9748-z
Gerhardt H, Betsholtz C (2003) Endothelial-pericyte interactions in angiogenesis. Cell Tissue Res 314(1):15–23. https://doi.org/10.1007/s00441-003-0745-x
Hampton D, Rhodes K, Zhao C, Franklin R, Fawcett J (2004) The responses of oligodendrocyte precursor cells, astrocytes and microglia to a cortical stab injury, in the brain. Neuroscience 127(4):813–820. https://doi.org/10.1016/j.neuroscience.2004.05.028
Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer cell 21(3):309–322. https://doi.org/10.1016/j.ccr.2012.02.022
Hayes BJ, Riehle KJ, Shimizu-Albergine M, Bauer RL, Hudkins KL, Johansson F, Yeh MM, Mahoney WM Jr, Yeung RS, Campbell JS (2014) Activation of platelet-derived growth factor receptor alpha contributes to liver fibrosis. PLoS ONE 9(3):e92,925. https://doi.org/10.1371/journal.pone.0092925
Heldin CH, Westermark B (1999) Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev 79(4):1283–1316. https://doi.org/10.1152/physrev.1999.79.4.1283
Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin CH, Westermark B, Nistér M (1992) Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res 52(11):3213–3219
Hoch RV, Soriano P (2003) Roles of PDGF in animal development. Development 130(20):4769–4784. https://doi.org/10.1242/dev.00721
Ingram JL, Rice AB, Geisenhoffer K, Madtes DK, Bonner JC (2004) Il-13 and il-1\(\beta \) promote lung fibroblast growth through coordinated up-regulation of PDGF-AA and PDGF-R\(\alpha \). FASEB J 18(10):1132–1134. https://doi.org/10.1096/fj.03-1492fje
Ivkovic S, Beadle C, Noticewala S, Massey SC, Swanson KR, Toro LN, Bresnick AR, Canoll P, Rosenfeld SS (2012) Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol Biol Cell 23(4):533–542. https://doi.org/10.1091/mbc.e11-01-0039
Juliano J, Gil O, Hawkins-Daarud A, Noticewala S, Rockne RC, Gallaher J, Massey SC, Sims PA, Anderson AR, Swanson KR et al (2018) Comparative dynamics of microglial and glioma cell motility at the infiltrative margin of brain tumours. J R Soc Interface 15(139):20170,582. https://doi.org/10.1098/rsif.2017.0582
Kakita A, Goldman JE (1999) Patterns and dynamics of SVZ cell migration in the postnatal forebrain: monitoring living progenitors in slice preparations. Neuron 23(3):461–472. https://doi.org/10.1016/S0896-6273(00)80800-4
Kohler N, Lipton A (1974) Platelets as a source of fibroblast growth-promoting activity. Exp Cell Res 87(2):297–301. https://doi.org/10.1016/0014-4827(74)90484-4
Levine JM, Reynolds R, Fawcett JW (2001) The oligodendrocyte precursor cell in health and disease. Trends Neurosci 24(1):39–47. https://doi.org/10.1016/S0166-2236(00)01691-X
Li H, Fan X, Houghton J (2007) Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biochem 101(4):805–815. https://doi.org/10.1002/jcb.21159
Lindmark G, Sundberg C, Glimelius B, Påhlma L, Rubin K, Gerdin B (1993) Stromal expression of platelet-derived growth factor beta-receptor and platelet-derived growth factor b-chain in colorectal cancer. Lab Investig J Tech Methods Pathol 69(6):682–689
Lopez K, Assanah M, Waziri A, Fusco D, Tannenbaum A, Linskey K, McKhann GM, Sisti MB, Bruce J, Canoll P (2008) Human glioma cells recruit and expand adult glial progenitors via paracrine platelet-derived growth factor signaling. Neurosurgery 62(6):1424–1425. https://doi.org/10.1227/01.neu.0000333529.85150.cf
Majesky MW, Reidy MA, Bowen-Pope DF, Hart CE, Wilcox JN, Schwartz SM (1990) Pdgf ligand and receptor gene expression during repair of arterial injury. J Cell Biol 111(5):2149–2158. https://doi.org/10.1083/jcb.111.5.2149
Majumdar K, Radotra BD, Vasishta RK, Pathak A (2009) Platelet-derived growth factor expression correlates with tumor grade and proliferative activity in human oligodendrogliomas. Surg Neurol 72(1):54–60. https://doi.org/10.1016/j.surneu.2008.10.001
Masliah E, Mallory M, Alford M, Deteresa R, Saitoh T (1995) PDGF is associated with neuronal and glial alterations of Alzheimer’s disease. Neurobiol Aging 16(4):549–556. https://doi.org/10.1016/0197-4580(95)00050-O
Massey SC (2017) Multi-scale modeling of paracrine PDGF-driven glioma growth and invasion. PhD thesis. https://digital.lib.washington.edu/researchworks/handle/1773/38058. Accessed November 05, 2018
Massey SC, Assanah MC, Lopez KA, Canoll P, Swanson KR (2012) Glial progenitor cell recruitment drives aggressive glioma growth: mathematical and experimental modelling. J R Soc Interface 9(73):1757–1766. https://doi.org/10.1098/rsif.2012.0030
Massey SC, Rockne RC, Hawkins-Daarud A, Gallaher J, Anderson ARA, Canoll P, Swanson KR (2018) Simulating PDGF-driven glioma growth and invasion in an anatomically accurate brain domain. Bull Math Biol 80(5):1292–1309. https://doi.org/10.1007/s11538-017-0312-3
Nakamura Y, Tanaka F, Yoshikawa Y, Mimori K, Inoue H, Yanaga K, Mori M (2008) PDGF-BB is a novel prognostic factor in colorectal cancer. Ann Surg Oncol 15(8):2129–2136. https://doi.org/10.1245/s10434-008-9943-9
Nunes MC, Roy NS, Keyoung HM, Goodman RR, McKhann G, Jiang L, Kang J, Nedergaard M, Goldman SA (2003) Identification and isolation of multipotential neural progenitor cells from the subcortical white matter of the adult human brain. Nat Med 9(4):439–447. https://doi.org/10.1038/nm837
Pringle N, Collarini EJ, Mosley MJ, Heldin CH, Westermark B, Richardson WD (1989) PDGF A chain homodimers drive proliferation of bipotential (O-2A) glial progenitor cells in the developing rat optic nerve. EMBO J 8(4):1049. https://doi.org/10.1002/j.1460-2075.1989.tb03472.x
Redwine JM, Armstrong RC (1998) In vivo proliferation of oligodendrocyte progenitors expressing PDGFAR during early remyelination. J Neurobiol 37(3):413–428. https://doi.org/10.1002/(SICI)1097-4695(19981115)37:3<413::AID-NEU7>3.0.CO;2-8
Rhodes K, Raivich G, Fawcett J (2006) The injury response of oligodendrocyte precursor cells is induced by platelets, macrophages and inflammation-associated cytokines. Neuroscience 140(1):87–100. https://doi.org/10.1016/j.neuroscience.2006.01.055
Ross R, Glomset J, Kariya B, Harker L (1974) A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro. Proc Natl Acad Sci 71(4):1207–1210. https://doi.org/10.1073/pnas.71.4.1207
Ross R, Raines EW, Bowen-Pope DF (1986) The biology of platelet-derived growth factor. Cell 46(2):155–169. https://doi.org/10.1016/0092-8674(86)90733-6
Roy NS, Wang S, Harrison-Restelli C, Benraiss A, Fraser RAR, Gravel M, Braun PE, Goldman SA (1999) Identification, isolation, and promoter-defined separation of mitotic oligodendrocyte progenitor cells from the adult human subcortical white matter. J Neurosci 19(22):9986–9995. https://doi.org/10.1523/JNEUROSCI.19-22-09986.1999
Scolding N, Franklin R, Stevens S, Heldin CH, Compston A, Newcombe J (1998) Oligodendrocyte progenitors are present in the normal adult human CNS and the lesions of multiple sclerosis. Brain 121(12):2221–2228. https://doi.org/10.1093/brain/121.12.2221
Suzuki SO, Goldman JE (2003) Multiple cell populations in the early postnatal subventricular zone take distinct migratory pathways: a dynamic study of glial and neuronal progenitor migration. J Neurosci 23(10):4240–4250. https://doi.org/10.1523/JNEUROSCI.23-10-04240.2003
Swan A, Hillen T, Bowman JC, Murtha AD (2018) A patient-specific anisotropic diffusion model for brain tumour spread. Bull Math Biol 80(5):1259–1291. https://doi.org/10.1007/s11538-017-0271-8
Swanson KR, Alvord EC Jr, Murray J (2000) A quantitative model for differential motility of gliomas in grey and white matter. Cell Prolif 33(5):317–329. https://doi.org/10.1046/j.1365-2184.2000.00177.x
Takayama S, Sasahara M, Iihara K, Handa J, Hazama F (1994) Platelet-derived growth factor b-chain-like immunoreactivity in injured rat brain. Brain Res 653(1):131–140. https://doi.org/10.1016/0006-8993(94)90381-6
Thorne RG, Hrabětová S, Nicholson C (2004) Diffusion of epidermal growth factor in rat brain extracellular space measured by integrative optical imaging. J Neurophysiol 92(6):3471–3481. https://doi.org/10.1152/jn.00352.2004
Tyagi V, Theobald J, Barger J, Bustoros M, Bayin NS, Modrek AS, Kader M, Anderer EG, Donahue B, Fatterpekar G et al (2016) Traumatic brain injury and subsequent glioblastoma development: review of the literature and case reports. Surg Neurol Int. https://doi.org/10.4103/2152-7806.189296
Ustach CV, Huang W, Conley-LaComb MK, Lin CY, Che M, Abrams J, Kim HRC (2010) A novel signaling axis of matriptase/PDGF-D/-PDGFR in human prostate cancer. Cancer Res 70(23):9631–9640. https://doi.org/10.1158/0008-5472.CAN-10-0511
van Heyningen P, Calver AR, Richardson WD (2001) Control of progenitor cell number by mitogen supply and demand. Curr Biol 11:232–241. https://doi.org/10.1016/S0960-9822(01)00075-6
van der Valk P, Lindeman J, Kamphorst W (1997) Growth factor profiles of human gliomas: do non-tumour cells contribute to tumour growth in glioma? Ann Oncol 8(10):1023–1029. https://doi.org/10.1023/A:1008265905505
Westermark B, Heldin CH, Nister M (1995) Platelet-derived growth factor in human glioma. Glia 15(3):257–63. https://doi.org/10.1002/glia.440150307
Wright JH, Johnson MM, Shimizu-Albergine M, Bauer RL, Hayes BJ, Surapisitchat J, Hudkins KL, Riehle KJ, Johnson SC, Yeh MM et al (2014) Paracrine activation of hepatic stellate cells in platelet-derived growth factor c transgenic mice: evidence for stromal induction of hepatocellular carcinoma. Int J Cancer 134(4):778–788. https://doi.org/10.1002/ijc.28421
Younger A, Wing K, Penner M, Cresswell M (2016) A study to evaluate the safety of platelet-derived growth factor for treatment of osteochondral defects of the talus. Knee Surg Sports Traumatol Arthrosc 24(4):1250–1258. https://doi.org/10.1007/s00167-015-3549-0
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-0781824, the James S. McDonnell Foundation Collaborative Activity Award #220020264, the National Institutes of Health (U01CA220378, U54CA210180, U54CA143970, U54CA193489, R01NS060752, R01CA16437, P01CA42045), the Ben and Catherine Ivy Foundation, the University of Washington Academic Pathology Fund, the James D. Murray Endowed Chair in the Nancy and Buster Alvord Brain Tumor Center at the University of Washington. The content is solely the responsibility of the authors. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation or any other supporting agencies. As always, KRS is eternally grateful to the unwavering support of Dr. E. C. “Buster” Alvord, Jr. (1923–2010); may this manuscript continue to honor his memory and foster his scientific legacy.
A Details of Derivation of \(p_0\)
A Details of Derivation of \(p_0\)
Recall from Sect. 2.3 that we have the following relationship:
which describes the increased activity of one condition of cells that has added exogenous PDGF relative to others that lack this added PDGF. Given that they are still active and have been injured during tissue removal, we assume that there is a shared baseline amount of PDGF, \(p_0\).
Expanding this relationship using the definition of R(p) in (6), we have:
Since \(\beta (EC_{50})\) is a constant, for notational simplicity, we set \(\gamma =\beta (EC_{50})\) throughout the following algebra. This lets us write
Then, we can cross multiply the denominators and expand the \(\beta \) terms:
Move all terms to one side for simplicity:
Now, multiply both sides by \(k_m + p_0\) and \(k_m+p_0+100\):
Expanding and then collecting by powers of \(p_0\):
Now, we can use the quadratic formula to find the roots of this equation, letting
for the formula
Recalling that we use parameter values \(k_m = 30\) ng/mL and \(EC_{50}=\sqrt{10}\), such that \(\gamma = \frac{EC_{50}}{k_m + EC_{50}} = \frac{\sqrt{10}}{30+\sqrt{10}} \), this computes to (in decimal approximation):
The latter of these does not make sense as a physical quantity, so we adopt the first as our approximate value for \(p_0\):
Rights and permissions
About this article
Cite this article
Massey, S.C., Hawkins-Daarud, A., Gallaher, J. et al. Lesion Dynamics Under Varying Paracrine PDGF Signaling in Brain Tissue. Bull Math Biol 81, 1645–1664 (2019). https://doi.org/10.1007/s11538-019-00587-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-019-00587-z