Abstract
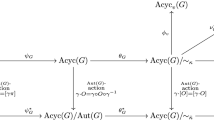
We present mathematical techniques for exhaustive studies of long-term dynamics of asynchronous biological system models. Specifically, we extend the notion of \(\kappa \)-equivalence developed for graph dynamical systems to support systematic analysis of all possible attractor configurations that can be generated when varying the asynchronous update order (Macauley and Mortveit in Nonlinearity 22(2):421, 2009). We extend earlier work by Veliz-Cuba and Stigler (J Comput Biol 18(6):783–794, 2011), Goles et al. (Bull Math Biol 75(6):939–966, 2013), and others by comparing long-term dynamics up to topological conjugation: rather than comparing the exact states and their transitions on attractors, we only compare the attractor structures. In general, obtaining this information is computationally intractable. Here, we adapt and apply combinatorial theory for dynamical systems from Macauley and Mortveit (Proc Am Math Soc 136(12):4157–4165, 2008. https://doi.org/10.1090/S0002-9939-09-09884-0; 2009; Electron J Comb 18:197, 2011a; Discret Contin Dyn Syst 4(6):1533–1541, 2011b. https://doi.org/10.3934/dcdss.2011.4.1533; Theor Comput Sci 504:26–37, 2013. https://doi.org/10.1016/j.tcs.2012.09.015; in: Isokawa T, Imai K, Matsui N, Peper F, Umeo H (eds) Cellular automata and discrete complex systems, 2014. https://doi.org/10.1007/978-3-319-18812-6_6) to develop computational methods that greatly reduce this computational cost. We give a detailed algorithm and apply it to (i) the lac operon model for Escherichia coli proposed by Veliz-Cuba and Stigler (2011), and (ii) the regulatory network involved in the control of the cell cycle and cell differentiation in the Caenorhabditis elegans vulva precursor cells proposed by Weinstein et al. (BMC Bioinform 16(1):1, 2015). In both cases, we uncover all possible limit cycle structures for these networks under sequential updates. Specifically, for the lac operon model, rather than examining all \(10! > 3.6 \cdot 10^6\) sequential update orders, we demonstrate that it is sufficient to consider 344 representative update orders, and, more notably, that these 344 representatives give rise to 4 distinct attractor structures. A similar analysis performed for the C. elegans model demonstrates that it has precisely 125 distinct attractor structures. We conclude with observations on the variety and distribution of the models’ attractor structures and use the results to discuss their robustness.







Similar content being viewed by others
References
Adiga A, Galyean H, Kuhlman CJ, Levet M, Mortveit HS, Wu S (2016) Activity in Boolean networks. Nat Comput 16:1–13
Aracena J, Goles E, Moreira A, Salinas L (2009) On the robustness of update schedules in Boolean networks. Biosystems 97(1):1–8
Coleman A (1989) Killing and the Coxeter transformation of Kac–Moody algebras. Invent Math 95(3):447–477
Demongeot J, Goles E, Morvan M, Noual M, Sené S (2010) Attraction basins as gauges of robustness against boundary conditions in biological complex systems. PLoS One 5(8):e11,793. https://doi.org/10.1371/journal.pone.0011793
Garg A, Di Cara A, Xenarios I, Mendoza L, De Micheli G (2008) Synchronous versus asynchronous modeling of gene regulatory networks. Bioinformatics 24(17):1917–1925
Goles E, Martínez S (2013) Neural and automata networks: dynamical behavior and applications, vol 58. Springer, Berlin
Goles E, Montalva M, Ruz GA (2013) Deconstruction and dynamical robustness of regulatory networks: application to the yeast cell cycle networks. Bull Math Biol 75(6):939–966
Hagberg A.A, Schult D.A, Swart P.J (2008) Exploring network structure, dynamics, and function using NetworkX. In: Proceedings of the 7th Python in Science Conference (SciPy2008). Pasadena, CA, USA, pp 11–15
Kitano H (2004) Biological robustness. Nat Rev Genet 5(11):826–837
Kuhlman CJ, Mortveit HS (2014) Attractor stability in nonuniform Boolean networks. Theor Comput Sci 559:20–33
Kuhlman CJ, Mortveit HS, Murrugarra D, Kumar V.A (2011) Bifurcations in Boolean networks. In: Discrete mathematics and theoretical computer science proceedings of the 17th international workshop on cellular automata and discrete complex systems (DMTCS Automata 2011), pp 29–46
Macauley M, Mortveit HS (2008) On enumeration of conjugacy classes of Coxeter elements. Proc Am Math Soc 136(12):4157–4165. https://doi.org/10.1090/S0002-9939-09-09884-0. arXiv:Math.CO/0711.1140
Macauley M, Mortveit HS (2009) Cycle equivalence of graph dynamical systems. Nonlinearity 22(2):421
Macauley M, Mortveit H.S (2011a) Posets from admissible Coxeter sequences. Electron J Comb 18(P197). Preprint: arXiv:math.DS/0910.4376
Macauley M, Mortveit HS (2011b) Update sequence stability in graph dynamical systems. Discret Contin Dyn Syst 4(6):1533–1541. https://doi.org/10.3934/dcdss.2011.4.1533 Preprint: arXiv:math.DS/0909.1723
Macauley M, Mortveit HS (2013) An atlas of limit set dynamics for asynchronous elementary cellular automata. Theor Comput Sci 504:26–37. Discrete Mathematical structures: from dynamics to complexity—DISCO 2011 24–26 November, 2011, Santiago, Chile. https://doi.org/10.1016/j.tcs.2012.09.015
Macauley M, Mortveit HS (2014) Cycle equivalence of finite dynamical systems containing symmetries. In: Isokawa T, Imai K, Matsui N, Peper F, Umeo H (eds) Cellular automata and discrete complex systems: 20th international workshop, AUTOMATA 2014, Himeji, Japan, July 7–9, 2014, Revised Selected Papers, Lecture Notes in Computer Science, vol 8996, pp 70–82 . https://doi.org/10.1007/978-3-319-18812-6_6
Montalva M, Ruz GA, Goles E (2014) Attraction basins in a lac operon model under different update schedules. In: ALIFE 14: The 14th conference on the synthesis and simulation of living systems 14:689–690
Müssel C, Hopfensitz M, Kestler HA (2010) Boolnet: an R package for generation, reconstruction and analysis of boolean networks. Bioinformatics 26(10):1378–1380
Řehůřek R, Sojka P (2010) Software framework for topic modelling with large corpora. In: Proceedings of the LREC 2010 workshop on new challenges for NLP frameworks. ELRA, Valletta, Malta, pp 45–50. http://is.muni.cz/publication/884893/en
Reidys C (1998) Acyclic orientations of random graphs. Adv Appl Math 21(2):181–192
Ruz GA, Goles E, Montalva M, Fogel GB (2014) Dynamical and topological robustness of the mammalian cell cycle network: a reverse engineering approach. Biosystems 115:23–32
Veliz-Cuba A, Stigler B (2011) Boolean models can explain bistability in the lac operon. J Comput Biol 18(6):783–794
Weinstein N, Ortiz-Gutiérrez E, Muñoz S, Rosenblueth DA, Álvarez-Buylla ER, Mendoza L (2015) A model of the regulatory network involved in the control of the cell cycle and cell differentiation in the caenorhabditis elegans vulva. BMC Bioinform 16(1):1
Acknowledgements
We thank our external collaborators and members of the Network Systems Science & Advanced Computing (NSSAC) division for their suggestions and comments. This work has been partially supported by DTRA Grant HDTRA1-11-1-0016.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mortveit, H.S., Pederson, R.D. Attractor Stability in Finite Asynchronous Biological System Models. Bull Math Biol 81, 1442–1460 (2019). https://doi.org/10.1007/s11538-018-00565-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-018-00565-x