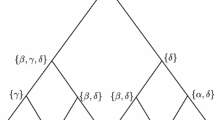
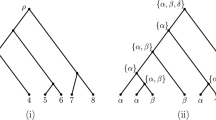
Abstract
One of the main aims in phylogenetics is the estimation of ancestral sequences based on present-day data like, for instance, DNA alignments. One way to estimate the data of the last common ancestor of a given set of species is to first reconstruct a phylogenetic tree with some tree inference method and then to use some method of ancestral state inference based on that tree. One of the best-known methods both for tree inference and for ancestral sequence inference is Maximum Parsimony (MP). In this manuscript, we focus on this method and on ancestral state inference for fully bifurcating trees. In particular, we investigate a conjecture published by Charleston and Steel in 1995 concerning the number of species which need to have a particular state, say a, at a particular site in order for MP to unambiguously return a as an estimate for the state of the last common ancestor. We prove the conjecture for all even numbers of character states, which is the most relevant case in biology. We also show that the conjecture does not hold in general for odd numbers of character states, but also present some positive results for this case.








Similar content being viewed by others
Notes
Note that inferring ancestral sequences with MP is sometimes referred to as ‘small parsimony’ problem. In contrast, the ‘big parsimony’ problem refers to inferring most parsimonious trees.
References
Cai W, Pei J, Grishin NV (2004) Reconstruction of ancestral protein sequences and its applications. BMC Evolut Biol 4:33
Felsenstein J (2004) Inferring phylogenies. Sinauer Associates, Inc., Sunderland
Fischer M, Liebscher V (2015) On the balance of unrooted trees. Preprint. arXiv:1510.07882
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Gascuel O, Steel M (2010) Inferring ancestral sequences in taxon-rich phylogenies. Math Biosci 227:125–153
Gascuel O, Steel M (2014) Predicting the ancestral character changes in a tree is typically easier than predicting the root state. Syst Biol 63:421–435
Goulden IP, Jackson DM (1983) Combinatorial enumeration. Wiley, New York
Griffith OW, Blackburn DG, Brandley MC, Van Dyke JU, Whittington CM, Thompson MB (2015) Ancestral state reconstructions require biological evidence to test evolutionary hypotheses: a case study examining the evolution of reproductive mode in squamate reptiles. J Exp Zool 324:493–503
Li G, Steel M, Zhang L (2008) More taxa are not necessarily better for the reconstruction of ancestral character states. Syst Biol 57:647–653
Liberles DA (ed) (2007) Ancestral sequence reconstruction. Oxford University Press, New York
Semple C, Steel M (2003) Phylogenetics. Oxford University Press, New York
Steel M, Charleston M (1995) Five surprising properties of parsimoniously colored trees. Bull Math Biol 57:367–375
Acknowledgements
We thank Mike Steel for bringing this topic to our attention. Moreover, we thank two anonymous reviewers for their helpful comments on an earlier version of this manuscript. The first author also thanks the Ernst-Moritz-Arndt-University Greifswald for the Landesgraduiertenförderung studentship, under which this work was conducted.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Proof
(Proof of Lemma 1) (i) Let A, B such that \(a \in A \cap B\) and \(|A| = |B|\). Then A could be transformed into B by renaming all states not element of \(A \cap B\). Then \(A=B\), and this yields \(f_{k,r}^A=f_{k,r}^B\).
(ii) Let \(k=0\). In this case, our tree consists of one leaf, which is at the same time the root. This vertex has to be assigned a to obtain \(X_\rho =\{a\}\). Hence \(f_{0,r}=1\) for all r.
(iii) Let \(k \ge 1\). Then
Therefore, \(a \in B\) and \(a \in C\) which results in \(f_{k,r} \ge 2\). \(\square \)
Proof
(Lemma 2) Let \(r= 2p\ge 2\), i.e. \(p\ge 1\).
-
1.
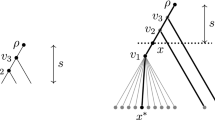
We start with the case \(k\le p\). We have \(f_{1,r}=2\) for all r (see Fig. 8). By Theorem 3, we know that \(f_{k,r}\) is monotonically increasing in k, and thus, \(2=f_{1,r} \le f_{p,r}\). We now use the standard decomposition for \(T_p\) in order to derive its two maximal pending rooted subtrees \(T_{p-1}\) with roots \(\rho _1\) and \(\rho _2\). We use the same construction as in the proof of Observation 1, which is depicted in Fig. 6. Thus, we can achieve \(X_{\rho _1}=A_p=\{a,c_1, \ldots , c_{p-1}\}\) and \(X_{\rho _2}=A_p^{'}=\{a,c_p, \ldots , c_{2p-2}\}\) by assigning a to one leaf in each subtree \(T_{p-1}\), respectively, where \(R=\{a,c_1,\ldots ,c_{2p-1}\}\), and such that all other subtrees use one state each which is unique to this subtree. Thus, the root of \(T_p\) will have the MP root state estimate \(A_p \cap A_p^{'}=\{a\}\). (Note that no leaf is assigned character state \(c_{2p-1}\); we do not even require all states. We will need this fact later.) We conclude \(f_{p,r}\le 2\), which together with \(f_{p,r}\ge 2\) as explained above completes \(f_{p,r}=2\). Together with Theorem 3, we achieve that \(f_{k,r}=2\) for all \(k=1,\ldots ,p\).
-
2.
Now consider the case \(k=p+1\). In this case, we have with (7)
$$\begin{aligned} f_{p+1,r}=f_{p+1,2p}&=\min \left\{ f_{p,2p}+f_{p,2p}^{A_{2p}},f_{p,2p}^{A_{2}}+f_{p,2p}^{A_{2p-1}},\ldots , f_{p,2p}^{A_{p}}+f_{p,2p}^{A_{p+1}} \right\} \\&=\min \left\{ f_{p,2p}+f_{p,2p}^{A_{2p}},f_{p-1,2p}+f_{p,2p}^{A_{2p-1}},\ldots , f_{1,2p}+f_{0,2p} \right\}&\text {by Theorem } 4 \\&=\min \left\{ 2+f_{p,2p}^{A_{2p}},2+f_{p,2p}^{A_{2p-1}},\ldots , 2+1 \right\}&\text {by Lemma 2, part 1 } \\&=3. \end{aligned}$$The latter equation is true because \(1 \le f_{p,2p}^{A_{2p}} \le f_{p,2p}^{A_{2p-1}} \le \cdots \le f_{p,2p}^{A_{p+1}}\) (at least one leaf has to be labelled a if a shall appear in the MP root state estimate).
-
3.
Now consider the case \(k=r\). We can proceed as above and assign \(X_{\rho _1}=A_p=\{a,c_1, \ldots , c_{p-1}\}\) to the first of the two \(T_{r-1}\) subtrees induced by the standard decomposition, and \(X_{\rho _2}=A_{p+1}^{'}=\{a,c_p, \ldots , c_{2p-1}\}\) to the other one, where again \(R=\{a,c_1,\ldots ,c_{2p-1}\}\). Then by (7), we can conclude \(f_{r,r}\le f_{r-1,r}^{A_p} + f_{r-1,r}^{A_{p+1}}\). Note that \(f_{r-1,r}^{A_p} + f_{r-1,r}^{A_{p+1}}= f_{p,r}+f_{p-1,r}\) by Theorem 4, so that \(f_{r,r}\le f_{p,r}+f_{p-1,r} = 2+2\), where the latter equation holds because of the first part of Lemma 2. Altogether, \(f_{r,r}\le 4\). By the monotonicity of Theorem 3, we obtain \(f_{k,r} \le 4\) for all \(p+1 < k \le r\). \(\square \)
Rights and permissions
About this article
Cite this article
Herbst, L., Fischer, M. Ancestral Sequence Reconstruction with Maximum Parsimony. Bull Math Biol 79, 2865–2886 (2017). https://doi.org/10.1007/s11538-017-0354-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-017-0354-6