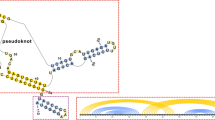
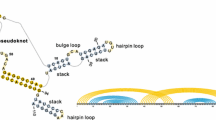
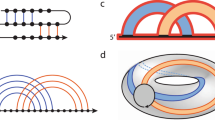
Abstract
In this paper, we present the asymptotic enumeration of RNA structures with pseudoknots. We develop a general framework for the computation of exponential growth rate and the asymptotic expansion for the numbers of k-noncrossing RNA structures. Our results are based on the generating function for the number of k-noncrossing RNA pseudoknot structures, \({\mathsf{S}}_{k}(n)\) , derived in Bull. Math. Biol. (2008), where k−1 denotes the maximal size of sets of mutually intersecting bonds. We prove a functional equation for the generating function \(\sum_{n\ge 0}{\mathsf{S}}_{k}(n)z^{n}\) and obtain for k=2 and k=3, the analytic continuation and singular expansions, respectively. It is implicit in our results that for arbitrary k singular expansions exist and via transfer theorems of analytic combinatorics, we obtain asymptotic expression for the coefficients. We explicitly derive the asymptotic expressions for 2- and 3-noncrossing RNA structures. Our main result is the derivation of the formula \({\mathsf{S}}_{3}(n)\sim \frac{10.4724\cdot4!}{n(n-1)\cdots(n-4)}(\frac{5+\sqrt{21}}{2})^{n}\) .
Similar content being viewed by others
References
Akutsu, T., 2000. Dynamic programming algorithms for RNA secondary structure prediction with pseudoknots. Discret. Appl. Math. 104, 45–62.
Chamorro, M., Parkin, N., Varmus, H.E., 1991. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl. Acad. Sci. USA 89, 713–717.
Chen, W.Y.C., Deng, E.Y.P., Du, R.R.X., Stanley, R.P., Yan, C.H., 2007. Crossings and nestings of matchings and partitions. Trans. Am. Math. Soc. 359, 1555–1575.
Flajolet, P., 1999. Singularity analysis and asymptotics of Bernoulli sums. Theor. Comput. Sci. 215(1–2), 371–381.
Flajolet, P., Fill, J.A., Kapur, N., 2005. Singularity analysis, Hadamard products, and tree recurrences. J. Comput. Appl. Math. 174, 271–313.
Flajolet, P., Grabiner, P., Kirschenhofer, P., Prodinger, H., Tichy, R.F., 1994. Mellin transforms and asymptotics: digital sums. Theor. Comput. Sci. 123, 291–314.
Gao, Z., Richmond, L.B., 1992. Central and local limit theorems applied to asymptotic enumeration. J. Appl. Comput. Anal. 41, 177–186.
Gessel, I.M., Zeilberger, D., 1992. Random walk in a Weyl chamber. Proc. Am. Math. Soc. 115, 27–31.
Haslinger, C., Stadler, P.F., 1999. RNA Structures with pseudo-knots. Bull. Math. Biol. 61, 437–467.
Hofacker, I.L., Schuster, P., Stadler, P.F., 1998. Combinatorics of RNA secondary structures. Discret. Appl. Math. 88, 207–237.
Howell, J.A., Smith, T.F., Waterman, M.S., 1980. Computation of generating functions for biological molecules. SIAM J. Appl. Math. 39, 119–133.
Jin, E.Y., Qin, J., Reidys, C.M., 2008. Combinatorics of RNA structures with pseudoknots. Bull. Math. Biol. 70, 45–67.
Konings, D.A.M., Gutell, R.R., 1995. A comparison of thermodynamic foldings with comparatively derived structures of 16s and 16s-like rRNAs. RNA 1, 559–574.
Lindstroem, B., 1973. On the vector representation of induced matroids. Bull. Lond. Math. Soc. 5, 85–90.
Loria, A., Pan, T., 1996. Domain structure of the ribozyme from eubacterial ribonuclease p. RNA 2, 551–563.
Lyngso, R., Pedersen, C., 1996. Pseudoknots in RNA secondary structures. In: H. Flyvbjerg, J. Hertz, M.H. Jensen, O.G. Mouritsen, K. Sneppen (Eds.), Physics of Biological Systems: From Molecules to Species. Springer, Berlin.
Mapping RNA Form and Function, 2005. Science 2, September 2005.
McCaskill, J.S., 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29, 1105–1119.
Odlyzko, A.M., 1992. Explicit tauberian estimates for functions with positive coefficients. J. Comput. Appl. Math. 41, 187–197.
Odlyzko, A.M., 1995. Handbook of Combinatorics. Elsevier, Amsterdam. Chapter 22.
Penner, R.C., Waterman, M.S., 1993. Spaces of RNA secondary structures. Adv. Math. 101, 31–49.
Popken, A., 1953. Asymptotic expansions from an algebraic standpoint. Indag. Math. 15, 131–143.
Rivas, E., Eddy, S., 1999. A dynamic programming algorithm for RNA structure prediction inclusing pseudoknots. J. Mol. Biol. 285, 2053–2068.
Schmitt, W.R., Waterman, M.S., 1994. Linear trees and RNA secondary structure. Discret. Appl. Math. 51, 317–323.
Tacker, M., Fontana, W., Stadler, P.F., Schuster, P., 1994. Statistics of RNA melting kinetics. Eur. Biophys. J. 23, 29–38.
Tacker, M., Stadler, P.F., Bauer, E.G., Hofacker, I.L., Schuster, P., 1996. Algorithm independent properties of RNA secondary structure predictions. Eur. Biophys. J. 25, 115–130.
Titchmarsh, E.C., 1939. The Theory of Functions. Oxford University Press, London.
Tuerk, C., MacDougal, S., Gold, L., 1992. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 89, 6988–6992.
Uemura, Y., Hasegawa, A., Kobayashi, S., Yokomori, T., 1999. Tree adjoining grammars for RNA structure prediction. Theor. Comput. Sci. 210, 277–303.
Waterman, M.S., 1978. Secondary structure of single-stranded nucleic acids. Adv. Math. I (suppl.) 1, 167–212.
Waterman, M.S., 1979. Combinatorics of RNA hairpins and cloverleafs. Stud. Appl. Math. 60, 91–96.
Waterman, M.S., Smith, T.F., 1986. Rapid dynamic programming algorithms for RNA secondary structure. Adv. Appl. Math. 7, 455–464.
Westhof, E., Jaeger, L., 1992. RNA pseudoknots. Curr. Opin. Struct. Biol. 2, 327–333.
Wong, R., Wyman, M., 1974. The method of Darboux. J. Approx. Theory 10, 159–171.
Zuker, M., Sankoff, D., 1984. RNA secondary structures and their prediction. Bull. Math. Biol. 46(4), 591–621.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jin, E.Y., Reidys, C.M. Asymptotic Enumeration of RNA Structures with Pseudoknots. Bull. Math. Biol. 70, 951–970 (2008). https://doi.org/10.1007/s11538-007-9265-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-007-9265-2