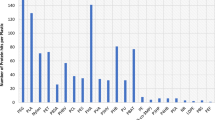
Abstract
The nitrogen cycle has been expanded with the recent discovery of Nitrospira strains that can conduct complete ammonium oxidation (commamox). Their importance in the nitrogen cycle within engineered ecosystems has not yet been analyzed. In this research, the community structure of the Bacteria domain of six full-scale activated sludge systems and three autotrophic nitrogen removal systems in the Netherlands and China has been investigated through tag-454-pyrosequencing. The phylogenetic analyses conducted in the present study showed that just a few of the Nitrospira sequences found in the bioreactors were comammox. Multivariate redundancy analysis of nitrifying genera showed an outcompetition of Nitrosomonas and non-comammox Nitrospira. Operational data from the bioreactors suggested that comammox could be favored at low temperature, low nitrogen substrate, and high dissolved oxygen. The non-ubiquity and low relative abundance of comammox in full-scale bioreactors suggested that this phylotype is not very relevant in the nitrogen cycle in wastewater treatment plants.







Similar content being viewed by others
References
Chao Y, Mao Y, Yu K, Zhang T (2016) Novel nitrifiers and comammox in a full-scale hybrid biofilm and activated sludge reactor revealed by metagenomics approach. Appl Microbiol Biotechnol 100:8225–8237. doi:10.1007/s00253-016-7655-9
Daims H, Lebedeva EV, Pjevac P et al (2015) Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi:10.1038/nature16461
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi:10.1093/nar/gkh340
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381
Gajewska M, Jóźwiakowski K, Ghrabi A, Masi F (2015) Impact of influent wastewater quality on nitrogen removal rates in multistage treatment wetlands. Environ Sci Pollut Res 22:12840–12848. doi:10.1007/s11356-014-3647-4
Ge S, Wang S, Yang X et al (2014) Detection of nitrifiers and evaluation of partial nitrification for wastewater treatment: a review. Chemosphere 140:85–98. doi:10.1016/j.chemosphere.2015.02.004
Gonzalez-Martinez A, Poyatos JM, Hontoria E, Osorio F (2011) Treatment of effluents polluted by nitrogen with new biological technologies based on autotrophic nitrification-denitrification processes. 74–84
Gonzalez-Martinez A, Rodriguez-Sanchez A, Rodelas B et al (2015a) 454-Pyrosequencing analysis of bacterial communities from autotrophic nitrogen removal bioreactors utilizing universal primers: effect of annealing temperature. Biomed Res Int. doi:10.1155/2015/892013
Gonzalez-Martinez A, Osorio F, Morillo JA et al (2015b) Comparison of bacterial diversity in full scale anammox bioreactors operated under different conditions. Biotechnol Prog n/a–n/a. doi:10.1002/btpr.2151
Gonzalez-Martinez A, Rodriguez-Sanchez A, Garcia-Ruiz MJ et al (2015c) Impact of methionine on a partial-nitritation biofilter. Environ Sci Pollut Res. doi:10.1007/s11356-015-5889-1
Gonzalez-Martinez A, Rodriguez-Sanchez A, Lotti T et al (2016a) Comparison of bacterial communities of conventional and A-stage activated sludge systems. Sci Rep 6:18786. doi:10.1038/srep18786
Gonzalez-Martinez A, Garcia-Ruiz MJ, Rodriguez-Sanchez A, Osorio F, Gonzalez-Lopez J (2016b) Archaeal and bacterial community dynamics and bioprocess performance of a bench-scale two-stage anaerobic digester. Appl Microbiol Biotechnol. doi:10.1007/s00253-016-7393-z
Gruber-Dorninger C, Pester M, Kitzinger K et al (2015) Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J 9:643–655. doi:10.1038/ismej.2014.156
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898. doi:10.1111/j.1462-2920.2010.02193.x
Jordaan K, Bezuidenhout CC (2015) Bacterial community composition of an urban river in the North West Province, South Africa, in relation to physico-chemical water quality. Environ Sci Pollut Res 1–13. doi:10.1007/s11356-015-5786-7
Lackner S, Gilbert EM, Vlaeminck SE, Joss A, Horn H, van Loosdrecht MCM (2014) Full-scale partial nitritation/anammox experiences—an application survey. Water Res 55:292–303. doi:10.1016/j.watres.2014.02.032
Nielsen PH, Mielczareck AT, Kragelund C et al (2010) A conceptual ecosystem model of microbial communities in enhanced biological phosphorous removal plants. Water Res 44:5070–5088. doi:10.1016/j.watres.2010.07.036
Pinto AJ, Marcus DN, Ijaz UZ, Bautista-de Iose Santos QM, Dick GJ, Raskin L (2015) Metagenomic evidence for the presence of Nitrospira-like bacteria in drinking water system. mSphere 1:e00054–e00015. doi:10.1128/mSphere.00054-15
Santoro AE (2016) The do-it-all nitrifier. Science 351:342–343. doi:10.1126/science.add9671
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/AEM.01541-09
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. doi:10.1371/journal.pone.0027310
Stöver BC, Müller KF (2010) TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. doi:10.1186/1471-2105-11-7
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Bio Evol 30(12):2725–2729. doi:10.1093/molbev/mst197
Tang HL, Chen H (2015) Nitrification at full-scale municipal wastewater treatment plants: evaluation of inhibition and bioaugmentation of nitrifiers. Bioresour Technol 190:76–81. doi:10.1016/j.biortech.2015.04.063
van Kessel MAHJ, Speth DR, Albertsen M et al (2015) Complete nitrification by a single microorganism. Nature 528:555–559. doi:10.1038/nature16459
Wang S, Peng Y, Ma B et al (2015) Anaerobic ammonium oxidation in traditional municipal wastewater treatment plants with low-strength ammonium loading: widespread but overlooked. Water Res 84:66–75. doi:10.1016/j.watres.2015.07.005
Wei YM, Wang JQ, Liu TT et al (2015) Bacterial communities of Beijing surface waters as revealed by 454 pyrosequencing of the 16S rRNA gene. Environ Sci Pollut Res 22:12605–12614. doi:10.1007/s11356-015-4534-3
Acknowledgments
The authors would like to acknowledge the support given by the Department of Civil and Environmental Engineering of the University of Aalto, Finland; the Institute of Water Research of the University of Granada, Spain; and the Department of Biotechnology of the Technical University of Delft, the Netherlands; as well, the authors would like to acknowledge the support provided by the personnel of the wastewater treatment plants of Amsterdam West, Breda, Dokhaven, Harnaschpolder, Kortenoord, Vianen, Apeldoorn, Meihua East, and Olburgen.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Gerald Thouand
Electronic supplementary material
Figure S1
Bayesian interference tree cladogram of all comammox candidate OTUs in the AS systems compared to Daims et al. 2015 phylogeny. (GIF 3045 kb)
Figure S2
Bayesian interference tree cladogram of all comammox candidate OTUs in the AS systems compared to van Kessel et al. 2015 phylogeny. (GIF 1432 kb)
Figure S3
Bayesian interference tree cladogram of all comammox candidate OTUs in the ANR systems compared to Daims et al. 2015 phylogeny. (GIF 2913 kb)
Figure S4
Bayesian interference tree cladogram of all comammox candidate OTUs in the ANR systems compared to van Kessel et al. 2015 phylogeny. (GIF 1671 kb)
Rights and permissions
About this article
Cite this article
Gonzalez-Martinez, A., Rodriguez-Sanchez, A., van Loosdrecht, M.C.M. et al. Detection of comammox bacteria in full-scale wastewater treatment bioreactors using tag-454-pyrosequencing. Environ Sci Pollut Res 23, 25501–25511 (2016). https://doi.org/10.1007/s11356-016-7914-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-7914-4