Abstract
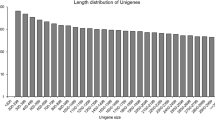
Tung oil, the major product of tung tree (Vernicia fordii) seeds, is one of the highest quality oils for industrial applications and has been considered for the production of biodiesel. Considering the poor agronomical traits of this crop, efforts have been made to breed tung trees for a higher fruit yield and oil property modification for biodiesel use or to engineer plants to produce a higher tung oil yield. However, these efforts have been hampered by a lack of molecular information, as there is no available genome and identified and characterized transcripts of this tree are scarce. Furthermore, there are still many knowledge gaps regarding tung oil biosynthesis. To provide a comprehensive and accurate foundation for molecular studies of tung tree, we present here a reference transcriptome dataset of mature tung seeds. A set of 43,081,927 reads were assembled into 47,585 unigenes. A homology search using blastx against the GenBank nonredundant protein database and the Swiss-Prot database resulted in the annotation of 96 and 81 % of these unigenes, respectively. We also systematically arranged a series of unigenes potentially associated with oil biosynthesis and degradation and examined the expression profile of a subset of those genes in samples from five different stages of seed development, providing a valuable source of genes and transcriptional information related to these pathways. This study represents the first large-scale transcriptome annotation of tung tree and will be useful in tung breeding for oil properties and other agronomical traits.







Similar content being viewed by others
References
Allen CAW, Watts KC, Ackman RG, Pegg MJ (1999) Predicting the viscosity of biodiesel fuels from their fatty acid ester composition. Fuel 78:1319–1326
Andre C, Haslam RP, Shanklin J (2012) Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. PNAS 109:10107–10112
Banik M, Duguid S, Cloutier S (2011) Transcript profiling and gene characterization of three fatty acid desaturase genes in high, moderate, and low linolenic acid genotypes of flax (Linum usitatissimum L.) and their role in linolenic acid accumulation. Genome 54:471–483
Brown K, Keeler W (2005) The history of tung oil. Wildland Weeds 9:4–24
Cahoon EB, Dietrich CR, Meyer K, Damude HG, Dyer JM, Kinney AJ (2006) Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochem 67:1166–1176
Cao H, Shockey JM, Klasson KT, Chapital DC, Mason CCB, Scheffler BE (2013) Developmental regulation of diacylglycerol acyltransferase family gene expression in tung tree tissues. Plos ONE 8:e76946
Chandran D, Sankararamasubramanian HM, Kumar MA, Parida A (2014) Differential expression analysis of transcripts related to oil metabolism in maturing seeds of Jatropha curcas L. Physiol Mol Biol Plants 20:181–190
Chen GQ, Turner C, He X, Nguyen T, McKeon TA, Laudencia-Chingcuanco D (2007) Expression profiles of genes involved in fatty acid and triacylglycerol synthesis in castor bean (Ricinus communis L.). Lipids 42:263–274
Chen Y, Zhou G, Wang Y, Xu L (2010) F-BOX and oleosin: additional target genes for future metabolic engineering in tung trees? Ind Crop Prod 32:684–686
Conesa S, Götz (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 1:12
Crawford JE, Guelbeogo WM, Sanou A, Traoré A, Vernick KD, Sagnon N, Lazzaro BP (2010) De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-Sseq technology. PLoS One 5:e14202
Dyer JM, Mullen RT (2005) Development and potential of genetically engineered oilseeds. Seed Sci Res 15:255–267
Dyer JM, Chapital DC, Kuan JCW, Mullen RT, Turner C, McKeon TA, Pepperman AB (2002) Molecular analysis of a bifunctional fatty acid conjugase/desaturase from tung implications for the evolution of plant fatty acid diversity. Plant Physiol 130:2027–2038
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Garg R, Patel RA, Khilesh A, Tyagi K, Jain MU (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18:53–63
Gruenheit N, Deusch O, Esser C, Becker M, Voelckel C, Lockhart PJ (2012) Cutoffs and k-mers: implications from a transcriptome study in allopolyploid plants. BMC Genomics 13:92
Gu K, Yi C, Tian D, Sangha JS, Hong Y, Yin Z (2012) Expression of fatty acid and lipid biosynthetic genes in developing endosperm of Jatropha curcas. Biotechnol Biofuels 5:47
Hu Y, Wu G, Cao Y, Wu Y, Xiao L, Li X, Lu C (2009) Breeding response of transcript profiling in developing seeds of Brassica napus. BMC Mol Biol 10:1–17
Jain RK, Coffey M, Lai K, Kumar A, MacKenzie SL (2000) Enhancement of seed oil content by expression of glycerol-3-phosphate acyltransferase genes. Biochem Soc Trans 28:958–961
Jaworski J, Cahoon EB (2003) Industrial oils from transgenic plants. Curr Opin Plant Biol 6:178–184
Jiang H, Wu P, Zhang S, Song C, Chen Y, Li M, Jia Y, Fang X, Chen F, Wu G (2012) Global Analysis of gene expression profiles in developing physic nut (Jatropha curcas L.) seeds. Plos One 7:e36522
Jolivet P, Boulard C, Bellamy A, Valot B, d’Andréa S, Zivy M, Nesi N, Chardot T (2011) Oil body proteins sequentially accumulate throughout seed development in Brassica napus. J Plant Physiol 168:2015-2020
Ke J, Wen TN, Nikolau BJ, Wurtele ES (2000) Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetyl-coenzyme a carboxylase. Plant Physiol 122:1057–1071
Knothe G (2005) Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Process Technol 86:1059–1070
Kroon JTM, Wei WX, Simon WJ, Slabas AR (2006) Identification and functional expression of a type 2 acyl-CoA: diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochem 67:2541–2549
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li R, Yu K, Hildebrand DF (2010) DGAT1, DGAT2 and PDAT expression in seeds and other tissues of epoxy and hydroxy fatty acid accumulating plants. Lipids 45:145–157
Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP et al (2013) Acyl-lipid metabolism. Arabidopsis Book 11:e0161
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆Ct Method. Methods 25:402–408
Lu CF, Wallis JG, Browse J (2007) An analysis of expressed sequence tags of developing castor endosperm using a full-length cDNA library. BMC Plant Biol 7:42
Maisonneuve S, Bessoule JJ, Lessire R, Delseny M, Roscoe TJ (2010) Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol 152:670–684
Meier S, Tzfadia O, Vallabhaneni R, Gehring C, Wurtzel ET et al (2011) A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. BMC Syst Biol 5:77
Muto M, Kubota C, Tanaka M, Satoh A, Matsumoto M, Yoshino T, Tanaka T (2013) Identification and functional analysis of delta-9 desaturase, a key enzyme in PUFA synthesis, isolated from the oleaginous diatom fistulifera. PlosOne 8:e73507
Nakkaew A, Chotigeat W, Eksomtramage T, Phongdara A (2008) Cloning and expression of a plastid-encoded subunit, beta-carboxyltransferase gene (accD) and a nuclear-encoded subunit, biotin carboxylase of acetyl-CoAcarboxylase from oil palm (Elaeis guineensis Jacq.). Plant Sci 175:497–504
Natarajan P, Parani M (2011) De novo assembly and transcriptome analysis of five major tissues of Jatropha curcas L. using GS FLX titanium platform of 454 pyrosequencing. BMC Genomics 12:191
Nguyen T, Shanklin J (2009) Altering arabidopsis oilseed composition by a combined antisense-hairpin RNAi gene suppression approach. J Am Oil Chem Soc 86:41–49
Park JY, Kim DK, Wang ZM, Lu P, Park SC, Lee JS (2008) Production and characterization of biodiesel from tung oil. Appl Biochem Biotechnol 148:109–117
Rajwade, AV, Kadoo NY, Borikar SP, Harsulkar AM, Ghorpade PB, Gupta VS (2014) Differential transcriptional activity of SAD, FAD2 and FAD3 desaturase genes in developing seeds of linseed contributes to varietal variation in a-linolenic acid content. Phytochem 98:41–53
Rupilinus W, Ahmad S (2007) Palm oil and palm kernel oil as raw materials for basic oleochemicals and biodiesel. Eur J Lipid Sci Technol 109:433–439
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864
Schmucki R, Berrera M, Küng E, Lee S, Thasler WE, Grüner S, Ebeling M, Cert U (2013) High throughput transcriptome analysis of lipid metabolism in Syrian hamster liver in absence of an annotated genome. Genomics 14:237
Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNAseq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092
Shang Q, Jiang W, Lu H, Liang B (2010) Properties of tung oil biodiesel and its blends with diesel. Bioresour Technol 101:826–828
Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK et al (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18:2294–2313
Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18:1961–1974
Surget-Groba Y, Montoya-Burgos JI (2010) Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res 20:1432–1440
Taylor DC, Zhang Y, Kumar A, Francis T, Giblin EM, Barton DL, Ferrie JR, Laroche A, Shah S, Zhu W, Snyder CL, Hall L, Rakow G, Harwood JL, Weselake RJ (2009) Molecular modification of triacylglycerol accumulation by over-expression of DGAT1 to produce canola with increased seed oil content under field conditions. Botany 87:533–543
Vanhercke T, Craig CW, Stymne S, Singh SP, Green AG (2013) Metabolic engineering of plant oils and waxes for use as industrial feedstocks. Plant Biotechnol J 11:197–210
Venturini L, Ferrarini A, Zenoni S, Tornielli GB, Fasoli M, Santo SD, Minio A, Buson G, Tononi P, Zago ED, Zamperin G, Bellin D, Pezzotti M, Delledonne M (2013) De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics 14:41
Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52:335–361
Wada M, Yasuno R, Jordan SW, Cronan JE, Wada H (2001) Lipoic acid metabolism in Arabidopsis thaliana: Cloning and characterization of a cDNA encoding lipoyltransferase. Plant Cell Physiol 42:650–656
Wang X-W, Luan J-B, Li J-M, Bao Y-Y, Zhang C-X, Liu S-S (2010) De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics 11:400
Xu R, Wang R, Liu A (2011) Expression profiles of genes involved in fatty acid and triacylglycerol synthesis in developing seeds of Jatropha (Jatropha curcas L.). Biomass Bioenergy 35:1683–1692
Xue J, Mao X, Yang Z, Wu Y, Jia X, Zhang L, Yue A, Wang J, Li R (2013) Expression of yeast acyl-CoA-D9 desaturase leads to accumulation of unusual monounsaturated fatty acids in soybean seeds. Biotechnol Lett 35:951–959
Yang W, Qi Y, Bi K, Fu J (2012) Toward understanding the genetic basis of adaptation to high-elevation life in poikilothermic species: a comparative transcriptomic analysis of two ranid frogs, Rana chensinensis and R. kukunoris. BMC Genomics 13:588
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Acknowledgments
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant number 559636/2009-1, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Financiadora de Projetos (FINEP), Embrapa, Petrobrás, and Ministério de Ciência e Tecnologia (MCT).
Data Archiving Statement
The RNA-Seq data and the transcriptome of tung tree seeds are currently been submitted to GenBank as Gene Expression Omnibus (GEO) data libraries. The accession numbers will be supplied once available.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. F. D. Dean
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
The five stages of fruit development used to obtain seeds for RT-qPCR analysis. S1: 20 days after flower opening (DAF); S2: 35 DAF; S3: 50 DAF; S4: 80 DAF; S5: 100 DAF. (GIF 21 kb)
Figure S2
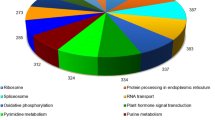
Distribution of tung contigs in metabolic pathways. The graphic representation of assigned metabolic pathways was obtained with the IPATH2 software (http://pathways.embl.de/ipath2) using the Blastall annotation. (PDF 402 kb)
Table S1
Primers used for RT-qPCR analysis. (XLSX 9 kb)
Table S2
Annotation of tung contigs according to the alignment against the GenBank non-redundant protein database (NR). The tung transcriptome contigs were utilized for homology searches against non-redundant sequences from NCBI (http://www.ncbi.nlm.nih.gov/) by applying the BLASTX program (e-value<1e-6). The bit score and e-value of the alignment, nucleotide and protein accession number, length, coding sequence (CDS) structure, and species with homologous sequences are presented. (XLSX 4555 kb)
Table S3
Annotation of tung contigs according to the alignment against the Swiss-Prot database. Contigs of the tung transcriptome were utilized for homology searches against protein sequences from the Swiss-Prot database (http://www.expasy.ch/sprot/) by applying the BLASTX program (e-value<1e-6). The bit score and e-value of the alignment, nucleotide and protein accession number, length, and species with homologous sequences are presented. (XLSX 3638 kb)
Table S4
KEGG annotation for tung contigs. KEGG pathway annotations were performed using the Blastall software against the KEGG databases (http://www.genome.jp/kegg/). (XLSX 1013 kb)
Table S5
The 50 most highly expressed contigs in tung seeds. The number of reads was calculated based on the alignment of the reads from RNA-Seq against the contigs. (XLSX 11 kb)
Table S6
Contigs related to oil biosynthesis in mature tung seeds. The BLASTX annotation against the Swiss-Prot and non-redundant sequence databases were used to search for sequences involved in oil biosynthesis. (XLSX 95 kb)
Table S7
Contigs related to oil breakdown in tung seeds. The BLASTX annotation against the Swiss-Prot and non-redundant sequence databases were used to search for sequences involved in oil breakdown. (XLSX 42 kb)
Rights and permissions
About this article
Cite this article
Galli, V., Guzman, F., Messias, R.S. et al. Transcriptome of tung tree mature seeds with an emphasis on lipid metabolism genes. Tree Genetics & Genomes 10, 1353–1367 (2014). https://doi.org/10.1007/s11295-014-0765-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0765-6