Abstract
We analyse the genetic variability in the yellow-necked mouse (Apodemus flavicollis) population in the city of Warsaw, Poland, and its surroundings - a species that has begun to occupy the city only in the last 30 years. We also compare the genetic variability of this species with corresponding data collected in the same time and areas for another species - the striped field mouse (Apodemus agrarius). The results indicate a gradual decrease in genetic diversity and increase in relatedness in the population of A. flavicollis from non-urban locations towards sites with the highest anthropopressure. The genetic structure was more pronounced in the 'recent invader' (A. flavicollis) than in the 'permanent inhabitant' (A. agrarius), which has a much longer city colonization history (more than 100 years). In general, FST was higher in A. flavicollis, which may indicate different and independent ways of city colonization by the species. The process by which urban areas are settled by a new, typically forest-dwelling species such as A. flavicollis, more 'sensitive' to the conditions of life in a city, probably includes not only successful events of penetration of the city by small groups of individuals (the founder effect), but also temporary extinctions of local urban populations of A. flavicollis or at least marked fluctuations in species population numbers. Suitably planned areas at the city borders could play an important role as 'gateways' through which individuals from non-urban populations could migrate into the city and join urban populations.
Similar content being viewed by others
Introduction
The process of urbanization is one of the most important factors affecting natural ecosystems at the beginning of the 21st century (Ricketts and Imhoff 2003; Goddard et al. 2010). Urban areas of the world are expected to absorb all the human population growth expected over the next four decades (United Nations 2011), and as urban populations expand, so do urbanized landscapes.
In general, the development of cities and other urbanized areas is the leading cause of habitat loss, threatening the existence of many wild species (Dearborn and Kark 2010). On the other hand, several studies have demonstrated that urban areas can harbour diverse habitats, simultaneously supporting high biodiversity (e.g. Muratet et al. 2007; Kadlec et al. 2008). Seeking a better understanding of how an urban environment can affect population processes in various species, molecular genetics methods have long been applied to analyse gene flow within urbanized landscapes and among urban landscapes and surrounding rural areas, as well as to estimate genetic variability in urban populations of wild species (e.g. Noël et al. 2007; Gardner-Santana et al. 2009; Gortat et al. 2013, 2015a; Munshi-South et al. 2013).
Although it is now obvious that the genetic after-effects of colonizing and/or living within an urban environment strongly depend on many factors, including the size and age of habitat patch, species mobility, and the characteristics of the matrix between patches, some general conclusions have been reached: (i) low-mobility species (e.g. amphibians), often related to a specific environment, such as water bodies, exhibit decreased genetic variability due to isolation and intensified genetic drift (Hitchings and Beebee 1998; Noël et al. 2007; Noël and Lapointe 2010; Mikulič and Pišút 2012; Munshi-South et al. 2013; Zak and Pehek 2013, but see also Saarikivi et al. 2013); (ii) mobile species able to cross environmental barriers seem to maintain a high level of genetic variability, probably avoiding complete isolation between patches of habitat within the city as well as between urban and rural areas (Desender et al. 2005; Gardner-Santana et al. 2009; Rutkowski et al. 2009; Bjӧrklund et al. 2010; Munshi-South and Kharchenko 2010); (iii) urban populations, usually fragmented into local subpopulations inhabiting small patches of suitable habitats (e.g. green spaces), exhibit pronounced genetic structure (Rubin et al. 2001; Wood and Pullin 2002; Wandeler et al. 2003; Desender et al. 2005; Gardner-Santana et al. 2009; Bjӧrklund et al. 2010; Delaney et al. 2010; Munshi-South and Kharchenko 2010; Vangestel et al. 2011; Gortat et al. 2013, 2015a; Munshi-South et al. 2013; Unfried et al. 2013).
Studies of urban populations have been focused mainly on species that have inhabited urban areas for a long time, or were even trapped by urban expansion and forced to exist in this transformed environment since its beginning (Chiappero et al. 2011; Munshi-South and Nagy 2014). Only a few studies, on the other hand, have looked at the distribution of genetic variability in species that have colonized cities recently. One such study investigated the population of foxes (Vulpes vulpes) in Zurich approx. 15 years after a significant increase in the number of animals within the city borders, and reported a small but significant genetic differentiation between urban and rural sampling sites, as well as slightly reduced genetic variability within urban populations, probably due to the founder effect (Wandeler et al. 2003). In another such study, the genetic variability of a breeding population of European kestrel (Falco tinnunculus) recently founded (in the 1970s) in the city of Warsaw was compared with rural populations and no differences were found, suggesting undisturbed gene flow between the relatively new urban population and birds from non-urban areas (Rutkowski et al. 2009).
The aim of our study was to analyse the distribution of genetic variability in the urban population of the yellow-necked mouse (Apodemus flavicollis) in Warsaw (Poland), which has long been numerous in the areas surrounding the city but appeared in Warsaw only very recently (approx. 30 years ago) and has colonized several patches within the city borders (Andrzejewski et al. 1978; Gliwicz 1980; Babińska-Werka and Malinowska 2008; Gortat et al. 2014). Moreover, given that another rodent species, the striped field mouse (A. agrarius), having a much longer city colonization history (more than 100 years), had previously been investigated in exactly the same locations (Gortat et al. 2015a), we were able to compare the results for the two species, using the same set of molecular markers. A. flavicollis and A. agrarius are very common species in Poland. A. flavicollis is a typical forest species while A. agrarius is habitat generalist and can occupy different kinds of green areas (Pucek 1984). We assumed that genetic processes, such as genetic drift, the founder effect, increased relatedness and genetic differentiation among local populations in the yellow-necked mouse population would be especially prominent in areas with a high degree of anthropopressure, i.e., located in the city centre, where anthropogenic barriers confine the rodents into small, spatially isolated green spaces. Moreover, because of fluctuations observed in small, isolated populations of A. flavicollis (Bujalska and Grüm 2005) we expected to see the genetic effects of population bottlenecks. We also assumed that the river cutting across the city, forming a natural barrier, would additionally limit the gene flow between the yellow-necked mouse populations settling areas on its opposite sides. Moreover, we anticipated that since the yellow-necked mouse has been inhabiting the city for a much shorter time than the striped field mouse, genetic differences between local populations of the first species would be much weaker.
Materials and methods
Study area and sample efforts
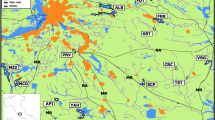
The study was carried out in the city of Warsaw (52°12'N, 21°02'E) and neighbouring non-urban areas (Fig. 1a). The city is located in the central part of Poland and its area covers 517 sq. km. The human population is estimated at 1.800.000. The Vistula River and a dense network of roads run through the city. Small green areas, as parks and gardens within the city are surrounded by dense urban buildings. Farther from the city centre, the urban infrastructure grows less developed, and so green areas situated at various distances from the city centre are exposed to different degrees of human pressure. Sudnik-Wójcikowska (1988), based on two main criteria – the intensity of human interference and the dominant vegetation type – distinguished five degrees of anthropopreassure, where ‘1’ means the lowest and ‘5’ the highest degree of human pressure. Particular degrees were defined as the percentage of the area potentially covered by vegetation: 1 zone (Z1) – natural vegetation covered 100% of the area; 2 zone (Z2) – almost natural vegetation covered more than 95% of the area; 3 zone (Z3) – seminatural vegetation covered not less than 95% of the area; 4 zone (Z4) – synanthropic vegetation covered 70-95% of the area; 5 zone (Z5) - synanthropic vegetation covered less than 70% of the area. Based on these criteria, Warsaw has been divided into a grid of squares (1.5 by 1.5 km) covering the area of the city and the degree of anthropopressure (anthropopressure zone) has been designated for each square in the grid (Sudnik-Wójcikowska 1988).
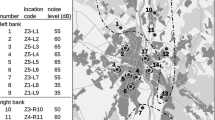
Scheme of the study area a and zones of anthropopressure b – in the city of Warsaw and neighbouring non-urban areas. Numbers on the maps denote particular locations where traps for small mammals were placed. Black points show locations where the number of A. flavicollis trapped was sufficient to include them in the genetic analysis. White points and cross points, respectively, denote locations where the number of individuals trapped was very small and locations where the mouse did not occur. Z1-Z5 – different zones of anthropopressure. NR – number of location. NL – noise level (dB)
We also assumed that city noise is an important stress-producing factor which can strongly restrict movements of rodents. For this reason the average daytime level of noise produced by city traffic expressed in decibels (dB) was used as an additional, direct indicator of anthropopressure (VIEP 2013) (Fig. 1b). Because in our study areas the correlation between the noise levels and the degree of anthropopressure proposed by Sudnik-Wójcikowska was very high and very strongly positive (r=0.9841; F=491.94, P<0.001) we decided to use both indicators (see also Gortat et al. 2015a).
Rodents were trapped at a total of 17 sites located on the right (R) and left (L) bank of the Vistula River (Fig 1a). Distances between particular locations within the city ranged from 2 km to about 20 km, and the overall surface area of the green spaces where the urban sites were located varied from about 27 ha to more than 600 ha. We assumed that the degree of anthropopressure at a particular trapping site was the same as that of the grid square in which it was located. In most cases, the degree of anthropopressure at the sites investigated decreased with growing distance from the centre of the city (r=0.9132; F=80.33, P<0.001). Four of them (#8, #9, #15, #16) were situated out of the city, two to the north and two to the south, on both sides of the river. All the non-urban locations were situated at the edge of large wooded areas and were exposed to the lowest degree of human pressure. The distance between the two farthest non-urban locations (#8 and #16) was about 50 km (Fig 1a).
Rodents were trapped at times of high densities of individuals (in September 2010 and 2011). At each site, a line running about 600 m was marked out, along which live traps for small mammals were placed. 30 trapping points were located along this line at each site, with 2 traps at each point. The trappings were carried out for 7 subsequent days in each location. Animals caught were individually marked and set free at the trapping site (prior approval for the study was obtained from the First Warsaw Local Ethics Committee for Animal Experimentation, no. 21/2010). Part of the ear of each animal was removed for molecular analysis.
Molecular analysis
Genomic DNA was extracted from the ear lobe samples of the yellow-necked mouse using the Genomic Mini Isolation Kit (A&A Biotechnology), according to the manufacturer’s instructions.
Ten microsatellite loci previously described by other authors (Gockel et al. 1997; Makova et al. 1998; Ohnishi et al. 1998; Harr et al. 2000) were analysed. All the markers (MSAf-3, MSAf-8, MSAf-22, CAA2A, GTTC4A, GACAD1A, GCATD7S, TNF, MSAA-5, As-7) were amplified in three independent multiplex reactions. PCR amplifications were performed in 12.5 μl reaction volume containing: 1 μl of template DNA, 6.25 μl 2x QIAGEN Multiplex PCR Master Mix (Qiagen), 1.25 μl Q-Solution 5x (Qiagen), 1.25 μl of 10x primer mix (containing each primer at 2 μM), 2.75 μl of RNase-free water (Qiagen). The reaction conditions were as follows: 94°C for 15 min, 30 cycles at 94°C for 30 s, 57°C for 3 min, 72°C for 1 min and a final extension at 68°C for 15 min. All reactions were carried out using a C-1000 Thermal Cycler (Bio-Rad).
Forward primers were labelled with one of the fluorescent WellRead labels (Sigma-Aldrich): Dye2; Dye3; Dye4. Negative PCR controls were always included for each set of reactions. No amplification product was found in any negative controls after electrophoresis in agarose gels and analysis in an automatic sequencer.
The length of the amplified fragments was measured using a CEQ8000 Beckman Coulter automatic sequencer. The data were analysed using Beckman Coulter Fragment Analysis Software.
Statistical analysis
Apodemus flavicollis
We assessed the observed heterozygosity (H O), unbiased expected heterozygosity (H E) (Nei and Roychoudhury 1974), and performed a probability test for deviation from Hardy-Weinberg equilibrium using Genepop on the Web version 4.0.10 (Raymond and Rousset 1995; Rousset 2008) for each locus within each of the sampling sites. Additionally, the fixation index (F IS) for each locus within each sampling site was calculated and its significance was tested under randomization procedure and Bonferroni correction for multiple comparison, using FSTAT version 2.9.3 (Goudet 2001). Although we found significant heterozygote deficiency in some loci, these deviations were not regularly observed across sampling sites – different loci exhibit heterozygote deficiency in different localizations. Moreover, significant F IS was found only in two cases: locus MSAf-3 at #1, and GTTC4A at #13.
Genetic variability, estimated based on microsatellite polymorphism, was evaluated at several levels. Firstly, we assessed allelic diversity (A), allelic richness (R; Petit et al. 1998), mean number of private alleles (P), private allelic richness (RP), observed heterozygosity (H O), unbiased expected heterozygosity (H E) (Nei and Roychoudhury 1974) and relatedness coefficient r (Queller and Goodnight 1989) separately for each sampling site. The fixation index (F IS) for each sampling site was also calculated and its significance was tested under randomization procedure and Bonferroni correction for multiple comparisons. These analyses were performed using GenAlEx version 6.0 (Paekall and Smouse 2001), FSTAT and HP-RARE (Kalinowski 2005). The genotypic linkage disequilibrium between all pairs of loci, as well as probability test for deviation from Hardy-Weinberg equilibrium, were evaluated using Genepop on the Web version 4.0.10 (Raymond and Rousset 1995; Rousset 2008) for each sampling site. Then the analyses were performed for each anthropopressure zone. The significance of differences between mean values of allelic richness (calculated across separate sampling sites within each anthropopressure zone), F IS, F ST, observed heterozygosity and relatedness in anthropopressure zones were tested using the permutation procedure as implemented in FSTAT. Similar comparisons were performed between river banks.
Effective population sizes for different populations were estimated from the sample data using LDNE (Waples and Do 2008). The program was run for each value of the lowest allele frequency used: 0.05, 0.02 and 0.01.
Evidence of recent effective population size reductions were investigated using Bottleneck 1.2 (Cornuet and Luikart 1996). Microsatellite data were tested under the stepwise mutation model (SMM) and Two-Phase Model (TPM) (Di Rienzo et al. 1994), with 95% SMM and variance of 12% (Piry et al. 1999). The significance of heterozygote excess was assessed by the Wilcoxon sign-rank test.
Genetic differentiation between sampling sites was estimated using F ST. Overall F ST (Weir and Cockerham 1984) for each anthropopressure zone and for each side of the river were obtained with FSTAT, as well as pairwise F ST among all sampling sites studied. The 95% confidence intervals for overall F ST were also estimated in FSTAT.
The relationship between the genetic distance and geographic distance was calculated according to the method proposed by Rousset (1997) and based on F ST/(1−F ST) index and natural logarithm of distance (ln m). In order to determine the relationship between the genetic and geographic distance of the population, the linear regression and Pearson product-moment correlation coefficient (r) were calculated, and the significance of the regression was evaluated by analysis of variance (ANOVA).
To obtain a picture of genetic differentiation among sites, we analysed our data using Principal Component Analysis (PCA) (PCA, Jombart et al. 2008), performed in Adegent 1.2-3 (Jombart 2008).
We also used the BAYESASS 3.03 program (Wilson and Rannala 2003), with default and adjusted settings, to assess recent migration rates and the direction taken by migration among sites (Wilson and Rannala 2003). We adjusted mixing parameters for allele frequencies (Δ A ), inbreeding coefficients (Δ F ) and migration rates (Δ M ) so that the acceptance rates were 38%, 45% and 44% respectively. MCMC was run for 10.000.000 steps, with 1.000.000 steps discarded as burn-in, and with sampling every 100th step. Convergence and mixing were inspected visually using Tracer v.1.5, with effective sample sizes greater than 100 assured for all the recorded parameters.
The STRUCTURE software package (version 2; Pritchard et al. 2000) was used to examine how well the predefined sampling sites corresponded to genetic groups (K). STRUCTURE was run 15 times for each user-defined K (1–10), with an initial burn-in of 50.000 and 100.000 iterations of the total data set. The admixture model of ancestry and the correlated model of allele frequencies were used. Sampling location was not used as prior information. Then we examined ΔK statistics that identify the largest change in the estimates of K produced by STRUCTURE, as ΔK may provide a more realistic estimation of K than those based on likelihoods (Evanno et al. 2005). As the method of Evanno et al. (2005) usually finds the uppermost level of genetic structure within a given dataset, the analysis was performed for the second time on genetic clusters, identified in analysis of the total data set. To visualize the STRUCTURE results, we used STRUCTURE HARVESTER (Earl and von Holdt 2011). Then we applied CLUMPP (Jakobsson and Rosenberg 2007) to average the multiple runs given by STRUCTURE and correct for label switching. The output from CLUMPP was visualized with DISTRUCT v 1.1 (Rosenberg 2004) to display the results.
Comparison of A. flavicollis and A. agrarius
Analysing two different rodent species in the same urbanized area, using microsatellite loci common for both species, gave us an opportunity to directly compare the results obtained. Hence, the genetic data about A. flavicollis obtained in the present study were directly compared to the results of microsatellite genotyping of A. agrarius, described in detail in another paper (Gortat et al. 2015a). We compared the levels of genetic variability and genetic differentiation found for the two species, sampled in exactly the same locations in Warsaw. In this analysis we included only microsatellite loci common for both species, namely: CAA2A, GTTC4A, GACAD1A, GCATD7S, TNF and MSAf-22. The significance of differences between mean values of allelic richness, F IS, F ST, observed heterozygosity and relatedness between species were tested using the permutation procedure as implemented in FSTAT.
Results
Genetic structure of Apodemus flavicollis
In total, we caught 295 individuals of A. flavicollis. The mouse was found to occupy 14 of the 17 locations covered by the study (Fig. 1). However, an insufficiently small number of individuals of the species (N<10) were caught at 4 sites, and so genetic analysis was performed on the basis of individuals caught at the 10 locations where number of individuals were higher and generally similar (20-26 mice) – with one exception (location #7, Fig. 1a), where 47 individuals were caught. Locations where we discovered no rodents or where the number of individuals was small were mostly situated in the areas with the highest degree of human pressure (zones Z4 and Z5, Fig. 1b).
We successfully amplified 10 microsatellites in 264 individuals of A. flavicollis. All loci were highly polymorphic in all investigated locations, with 10–22 alleles per locus. The highest allelic diversity and allelic richness were found in non-urban locations (Z1, Table 1). The lowest number of microsatellite alleles was identified in location #4, grouped in zone 5 of anthropopressure, on the left side of the Vistula River (Table 1). Allelic richness, calculated for locations from Z1 (R = 8.463), was significantly higher (two-sided test, 1000 permutations, P = 0.002) than allelic richness in pooled urban locations (Z3–Z5, R = 6.484). Comparing locations from particular zones, we found only significant differences (P= 0.002) in R between Z1 (R= 8.463) and Z4 (R = 6.023). However, in these pairwise comparisons, allelic richness was always slightly higher in zones with lower anthropopressure level, with the exception of #14, which showed a higher mean number of alleles than each of the locations from Z4. There was also no significant difference in R between sites from the left and right river bank.
There was no clear relation between the number of private alleles and zones of anthropopressure (Table 1). The highest mean number of private alleles was found in #1 and #9, and the lowest in #8 and #4. In the remaining zones, private alleles and private allelic richness were similar, ranging from 0.10 to 0.30.
Heterozygosity was observed to be below 0.70 only at three locations in the zones of highest anthropopressure: two from Z4 and one from Z5 (Table 1). The highest values were found in #9 and #16. Heterozygosity was not significantly different either between non-urban and urban locations (H O = 0.756 and 0.707 for non-urban and urban, respectively, two-sided test, 1000 permutations, P>0.05), or in any other comparisons among zones and river sides. Similarly, we did not find significant differences in F IS values. Considering particular locations, values of F IS were generally low and insignificant after the Bonferroni correction, except #14 and #13, respectively, from zones Z5 and Z4 on the right bank of the Vistula River. Among the 10 investigated locations, 5 were not in Hardy-Weinberg equilibrium (Table 1).
We found a clear relation between anthropopressure and effective population size N e (Table 1). The highest values of N e were found in non-urban locations; however, similar values were also found in adjacent urban areas from Z3 and Z4 (#7 and #12). The lowest N e was found in sites with the highest anthropopressure on both sides of the river, as well as in #13 from Z4.
The relatedness coefficient was significantly lower in non-urban locations than in urban ones (r = 0.176 and 0.254 for Z1 and Z3-Z5 respectively, P = 0.045) and significantly higher in Z5 than in Z1 (0.309 and 0.176, respectively, P = 0.001). When relatedness was compared among individuals in particular locations (Fig. 2), a clear pattern was observed, indicating a statistically significant gradual increase in mean relatedness as anthropopressure increased (r=0.9295; F=57.18, P<0.0003). In all locations, mean relatedness among individuals was higher than value expected for random mating.
We found no evidence of a recent bottleneck in non-urban and urban locations, independently of anthropopressure zone.
The overall genetic differentiation was high (F ST = 0.134, 95% CI 0.118–0.149). Only 13% of pairwise comparisons indicated F ST lower than 0.10. The higher percentage of values (58%) ranged from 0.101 to 0.15. Values over 0.20, suggesting strongly reduced gene flow, were found only in four cases (9% of all comparisons), all for pairwise comparison between #4 and locations on the right side of the river (#12, #13, #14, #16 Table 2, Fig. 1). The smallest values of pairwise differentiation (F ST, Table 2) were found for comparisons of non-urban locations (Z1) and the nearest urban locations on the same river side (#9 - #7, #8 - #1, #16 - #13 – Fig. 1). Small genetic differentiation was also found for comparison of locations from Z1 – only three comparisons exceeded the value of 0.10, corresponding to sampling sites located on opposite river banks and sides of the city (#9 - #15, #8 - #16, #9 - #16 – Fig. 1.). Genetic differentiation (F ST) was significantly lower in non-urban (Z1) than in all urban locations (0.100 and 0.149, respectively, P = 0.045) and in Z1 than Z4 (0.100 and 0.188, respectively, P = 0.004). There were no significant differences between Z3 and Z4, and Z1 and Z3, or between the left and right river bank.
The relationship between genetic distance (F ST/1- F ST) and geographic distance (ln m) calculated for all locations was negative but not statistically significant (r = -0.2665, ANOVA: F = 3.286329, P=0.07) (Fig. 3). The highest values of genetic distance were found in 3 locations situated close to each other in zones of the highest degree of anthropopressure (Z4-Z5), the smallest between the most geographically distant locations situated on the opposite sides of the city from Z1 (Figs. 1 and 3).
The relationship between genetic distance [F ST/(1−F ST)] and natural logarithm of geographic distance (ln m) calculated for all locations. Triangles and white dots represent pair of locations from highest (Z4-Z5) and lowest zones (Z1-Z3) of anthropopressure, respectively. Black dots show remaining pairs of locations
We found a low level of gene flow among the locations investigated (additional data are given in Online Resource 1). In the majority of cases, the ratio of resident individuals exceeded 85%. Only in #9 and #16 from Z1 did we find the fraction of migrants per generation to be higher than 15%. In both of these non-urban locations, the migrants derived from zones with higher anthropopressure, suggesting that gene flow occurred from urban locations into the nearest/neighbouring non-urban locations.
PCA (Fig. 4) suggested the presence of three groups of locations. The first, with negative eigenvalue scores, consists of two non-urban sampling sites located to the north of Warsaw on both river banks (#15 and #8 from Z1) plus one Z3 location (#1), also to the north of the city centre. The second group consists of three locations from the left river bank, located southwards of the city centre (#4, #7, #9). The third group consists of four sampling sites in right-bank Warsaw, including three urban sites (#12, #14, #13) plus an non-urban one (#16), located to the south of the city centre.
The STRUCTURE results (analysis of the total data set) indicated a gradual increase in mean likelihood from K=1 to K=9 (Fig. 5a). ΔK indicated the presence of two genetic clusters (Fig. 5b), and analysis of individual probability of ancestry suggested that these clusters strictly correspond to opposite river banks (Fig. 5c). However, in the non-urban sampling locations from the left side of the river, a small portion of individuals had higher probability of ancestry from the right-bank cluster. In #14, one individual of mixed origin from both clusters was indicated.
Estimated likelihoods, ln P(D), of each number of inferred genetic clusters a, the corresponding ΔK curves as a function of K b in population of A. flavicollis from Warsaw and neighbouring rural areas, and Bayesian assignment of individuals to genetic groups, indicated by ΔK c. Each bar represents the estimated posterior probability of each individual belonging to each of the two inferred clusters. Solid black lines define the boundaries between the local populations used in the analysis. Results for 10 populations, analysed altogether
The analysis performed on data from the left bank of the river (Fig. 6a, b and c) pointed to a division into three or five genetic clusters (Fig. 6b). In the three-clusters scenario, the population from the left bank non-urban sites from south and north were combined into common clusters with adjacent urban areas (Fig. 6c). Individuals assigned to northern cluster (#1 and #8) were additionally found in #9 (southward of Warsaw). Individuals from a location in the centre of the city (#4 from Z5) were assigned to a separate cluster (Fig. 6c). In the five-clusters scenario, in turn, genetic groups correspond to particular locations, with some signs of admixture between #8 and #1, and between #7 and #9 (Fig. 6c).
Estimated likelihoods, ln P(D), of each number of inferred genetic clusters a, the corresponding ΔK curves as a function of K b in population of A. flavicollis from Warsaw and neighbouring rural areas, and Bayesian assignment of individuals to genetic groups, indicated by ΔK c. Each bar represents the estimated posterior probability of each individual belonging to each of the two inferred clusters. Solid black lines define the boundaries between the local populations used in the analysis. Results for populations located on the left bank of the river
The analysis performed on the data from the right bank of the river (Fig. 7a, b and c) indicated the presence of four or five genetic groups. In the four-clusters scenario, two sites, located southward of the centre of Warsaw, were combined into single group. In the five-cluster scenario, genetic groups corresponded to particular locations (Fig. 7c). In both cases, in non-urban sites, we found individuals from the other non-urban clusters. Additionally, the results suggested that #13 site has some individuals from #14 (Fig. 7c).
Estimated likelihoods, ln P(D), of each number of inferred genetic clusters a, the corresponding ΔK curves as a function of K b in population of A. flavicollis from Warsaw and neighbouring rural areas, and Bayesian assignment of individuals to genetic groups, indicated by ΔK c. Each bar represents the estimated posterior probability of each individual belonging to each of the two inferred clusters. Solid black lines define the boundaries between the local populations used in the analysis. Results for populations located on the right bank of the river
Comparison of the genetic structure of A. flavicollis and A. agrarius
The analysis based on microsatellites limited to loci successfully amplified in both species (data for A. agrarius from Gortat et al. 2015a) indicated higher genetic variability in A. flavicollis in the area involved in the study. Allelic richness in 6 microsatellites was significantly higher in A. flavicollis than in A. agrarius (Table 3, ‘All sampling sites’). Similarly, higher heterozygosity was observed in A. flavicollis. The difference in genetic variability between species was identified for non-urban sampling sites (Table 3, ‘Non-urban sampling sites’), as well as for urban locations from Z3–Z5 (Table 3, ‘Urban sampling sites‘), but a significant difference was observed only in the case of heterozygosity. The heterozygosity observed was significantly higher in A. flavicollis both on the left and right side of the river, while the allelic richness was significantly higher only on the right bank of the river (Table 3, ‘All sampling sites (left bank)’ and ‘(right bank)’). In non-urban locations (Z1), differences in the genetic variability were significant on both sides of the river (Table 3, ‘Non-urban sampling sites (left bank)‘ and ‘(right bank)’), while in the urban (Z3–Z5) locations, significant differences in R and H O were found only on the right side of the river (Table 3, ‘Urban sampling sites (left bank)’ and ‘(right bank)’). If locations within each species were not divided into urban and non-urban, the inbreeding coefficient F IS was significantly different between A. flavicollis and A. agrarius only on the left bank of the Vistula River, with lower value for the former species (Table 3, ‘All sampling sites (left bank)’). There were no significant differences in F IS when comparing non-urban sites, but the value was always lower for A. flavicollis (Table 3, ‘Non-urban sampling sites’). In the case of urban sampling sites, a significant difference in F IS was found only on the right bank of the river, with the lower value for A. agrarius.
Except for comparison of urban sampling sites from the right side of the Vistula River, the relatedness coefficient r was always clearly higher in A. flavicollis, although significant differences were only found for all sampling sites, all sampling sites on the left bank, and in comparison of all non-urban sampling sites (Table 3).
A direct comparison of the genetic differentiation between species was performed based on overall F ST values. We found that the genetic differentiation was more pronounced in A. flavicollis. Significantly higher F ST values for A. flavicollis than A. agrarius, indicating more restricted gene flow in the former species, were found in the case of comparison of all sampling sites and all sampling sites from the left bank of the river (Table 3). In general, F ST was higher in A. flavicollis, independently of sampling site grouping, except for urban sites from the right bank of the Vistula River, where values for both species were very similar (Table 3).
Discussion
The process of colonization of a new area by a species is usually interlinked with one-time or serial events, involving migrations of a small number of individuals. In theory, the establishment of a population within a newly colonized area subsequently leads to a reduction in genetic variability due to the founder effect and the appearance of pronounced local genetic differentiation (Chakraborty and Nei 1977; Avise 1994). However, such a founder effect is significantly less intensive when colonization involves multiple source populations, which exchange gene pools after immigration into the new area (Slatkin 1977; Whitlock and McCauley 1990). Moreover, when a newly established population starts to expand rapidly and is continuously connected with source populations by a gene flow, the effect of colonization on genetic diversity of a founding population may be limited (McCauley 1991). This is evident in species with an ability for long-range dispersal or in cases when the area colonized borders on the territory of the source population (Dlugosch and Parker 2008).
Based on our genetic study of A. flavicollis in Warsaw, we found that, in a species colonizing the urban environment, genetic diversity gradually decreases from non-urban sites towards locations within the central area of the city. In this respect, our data confirmed the previous results for another rodent species in Warsaw, A. agrarius (Gortat et al. 2015a). Such a pattern occurs when groups of rodents become isolated within patches of habitat, surrounded by unsuitable areas, and subsequently, isolation induces genetic drift, decreasing the genetic variability. Indeed, in A. flavicollis, we found very low gene flow among the investigated sites. A decrease in genetic diversity was clearly interlinked with an increase in relatedness and smaller effective size of populations. Similarly, this could be an after-effect of intensified isolation, but could also be interlinked with colonization of the urban habitat by small groups of animals from neighbouring non-urban locations, as ‘stepping stones’, with simultaneous founder effects. Such a pattern of expansion usually leads to reduced genetic diversity in individuals inhabiting newly colonized areas as compared to source population (Dlugosch and Parker 2008). Indeed, we found higher genetic diversity in non-urban than pooled urban sampling sites. This corresponds to the history of A. flavicollis in Warsaw and the surroundings of the city: sites within the urban environment have been colonized very recently, whereas the species had inhabited natural (forest) areas around Warsaw long before colonizing the city (Andrzejewski et al. 1978; Gliwicz 1980; Babińska-Werka and Malinowska 2008). Interestingly, significant differences were confirmed only in terms of microsatellite alleles, but not in the comparison of heterozygosities. As predicted by theory, genetic drift in isolated populations acts more efficiently on the number of alleles than the level of heterozygosity (Nei et al. 1975). Hence, we can assume that 30 years is too short a period to significantly reduce the level of heterozygosity, even in the most isolated locations from Z5. A similar pattern was also found in A. agrarius, but in this species, significant differences between Z5 and less isolated locations were indicated also for heterozygosity (Gortat et al. 2015a). Obviously, this reflects the much longer (over one hundred year) history of the urban population of A. agrarius in Warsaw.
Genetic differentiation of A. flavicollis within the city area was high and significantly higher than the differentiation among non-urban locations, despite the smaller geographic distance separating the urban sites. The existence of sub-structuring within the urbanized landscape reflects the isolation of suitable habitat patches by artificial barriers, such as roads, heavily built-up areas, etc. In the investigated species, these barriers seem to be very efficient, as supported by the analysis of gene flow (BayesAss). Such a pattern has been suggested for many other species, including rodents (e.g. Gardner-Santana et al. 2009; Munshi-South and Kharchenko 2010; Munshi-South et al. 2013; Gortat et al. 2013, 2015a). However, given the very recent colonization of Warsaw by A. flavicollis, a clearly pronounced genetic structure was rather unexpected. Similar results were obtained during a study of recently founded urban population of Norway rat (Rattus norvegicus) (Kajdacsi et al. 2013), where they were explained more in terms of rapid population turnovers occurring in urban rodents than the social structure of the species.
According to Bujalska and Grüm (2005), the distinctive social and spatial organization of populations of the yellow-necked mouse, involving a lack of territorial behaviour in mature females, may be a cause for the frequent uncontrolled peaks and subsequent dramatic decreases of population numbers. This leads to poor winter survival rates and frequent very low concentrations in spring, which may result in extinction events of species populations (Bujalska and Grüm 2006, 2008) and, simultaneously, in saturation dispersal of groups of individuals during periods of high population numbers (Lidicker 1975), leading to colonization of new sites. In their long-term study of a population of that species settling a lake island with area of 4.5 ha, Bujalska and Grüm (2008) noted several extinction events, followed by recolonization. Gortat et al. (2015b) also found dramatic seasonal “booms and busts” in the population size and genetic diversity in a population of yellow-necked mouse inhabiting an island of similar size. It can therefore be expected that, in a city environment as examined herein, repeated extinction and recolonization events of local (island) populations of the yellow-necked mouse will take place, and the existence of permanent populations of the species within a city will be possible only in green spaces such as parks and woods of suitably large area, much greater than 4.5 ha. Simultaneously, frequent decreases in numbers of local populations of the species, leading to random loss of genetic diversity (the bottleneck effect) and to the recolonization, by small groups of dispersers, of habitat patches in which local populations went extinct (the founder effect) will contribute to a constant level of genetic variability and strongly pronounced genetic structure of the city population of the species. In the investigated locations we did not confirm genetic after-effects of population bottlenecks. This suggests that the sampled populations are rather stable, as is further supported by the clear genetic structure. Moreover, it can be expected that the occurrence of multiple paternity in A. flavicollis could partly compensate for the loss of genetic diversity by avoidance of inbreeding and increasing of offspring survival (Gryczyńska-Siemiątkowska et al. 2008). In contrast, A. agrarius was found in all of the 17 locations studied (Gortat et al. 2015a). This species is characterized by critical density values above which sexual maturity of females is delayed and therefore only a limited number of mature females is observed in these populations (Bujalska 1981). This adaptation, as well as multiple paternity which was also found in A. agrarius (Baker et al. 1999), may be the important factors supporting the existence of local populations in the cities where green areas are usually small and spatially isolated.
We found a few possible patterns explaining the genetic differentiation among the investigated subpopulations of A. flavicollis. Firstly, the results of the STRUCTURE analysis indicate the existence of two genetic groups (clusters) in the studied population of A. flavicollis, occurring on opposite sides of the river (Fig. 5). This is undoubtedly a result of the population being divided by an effective natural barrier preventing gene flow, such as a river may pose for small rodents (Aars et al. 1998; Gerlach and Musolf 2000; Booth et al. 2009; Gortat et al. 2015a). The effect is much stronger within the city borders than outside, as is shown by the PCA and STRUCTURE results. The river probably has a similar effect in urban and non-urban areas but in tandem with the impact of additional barriers (as roads, buildings, unsuitable habitats, etc.) the effect of genetic isolation of local populations is higher in the city. A similar gene flow pattern in the city population of rodents in Warsaw was found by Gortat et al. (2015a) for the striped field mouse.
Secondly, genetic differentiation was the smallest between a given non-urban location and the closest urban location, suggesting ongoing gene flow between these two areas. As indicated by BayesAss and STRUCTURE, gene flow occurs more from urban locations to the non-urban environment than in the opposite direction. This suggests that areas of the city with a lower level of anthropopressure (Z3) constitute a ‘tempting’ and suitable environment for rodents; however, during dispersal, individuals tend to migrate to zones of different anthropopressure level and then penetrate the urban environment more deeply. Most probably, the direction of migration was affected by the presence of barriers and larger areas of unsuitable habitat towards zone 4. This, in turn, suggests that migrations towards the city centre are rather scarce, resulting in isolation and reduced genetic diversity of groups of A. flavicollis in the highly transformed urban habitat.
Our analysis indicated that within the investigated area, spreading over the city of Warsaw and its environs, the genetic structure is much more pronounced in A. flavicollis than in A. agrarius. Although comparison between species was based on a relatively small number of microsatellites, we suggest that the results (direct comparison of F ST values, STRUCTURE) did not confirm the hypothesis that the much shorter presence of the yellow-necked mouse in the city, as compared to that of the striped field mouse, should entail smaller genetic differences between local populations of the former species. It may be speculated that the reason for this may lie in the city having been colonized in different and independent ways by the yellow-necked mouse. Unlike the yellow-necked mouse, the striped field mouse has been present in Warsaw for more than 100 years (Walecki 1881; Suminski 1922) and it forms stable local populations in many green areas in the city, including ones in the highest anthropopressure zone (Gortat et al. 2014). The genetic structure of the urban population of this species is most strongly expressed in the central part of the city, where particular local populations are strictly isolated from one another by city infrastructure (Gortat et al. 2015a). In parts of the city where anthropopressure is lower, and in non-urban areas, the genetic structure of the species is not so distinct.
The previous investigation of A. agrarius population from Warsaw indicated that, in left-bank Warsaw (a more transformed environment), non-urban populations were connected by gene flow only with the neighbouring locations, while more central sampling sites were highly differentiated due to isolation and genetic drift (Gortat et al. 2015a). In the present study, we have confirmed this pattern also in A. falvicollis. PCA indicated the presence of two genetic groups on the left side of the Vistula River (northern and southern), separating the city (Fig. 4). However, on the right side of the river (less transformed and with several large patches of green space, often connected with each other by green corridors) we found a contrasting pattern of grouping. Whereas in A. agrarius the northern non-urban population exchanges genes with urban locations as far away as the southern border of the city (Gortat et al. 2015a), in A. flavicollis the right-side locations seem to be better connected with the genetic pool of the southern non-urban population (Fig. 4). The differences between the two species as to gene exchange between their local populations may be connected with their different habitat requirements. The striped field mouse is quicker to move through green spaces irrespectively of their afforestation rate. For the yellow-necked mouse, in turn, the prevalence of woodlands (such as in the areas on the right bank of Vistula to the south of the city) may be crucial for attempts to settle new habitat patches. A. flavicollis, as a forest species, is behaviourally less inclined to migration through open areas, and requires more ‘comfortable’ corridors which are rear in heavily built-up areas. Another factor preventing it from expansion into urban areas, could be its diet, composed preferably of large tree seeds (Pucek 1984) often not available in sufficient quantities in the cities. We do not know why A. flavicollis has only recently started to colonize the city. The species has been present in areas surrounding Warsaw for a long time and it might be suggested that the processes of urban growth and development could be one of the possible reasons explaining its recent invasion of urban areas.
Both species of mice may be treated as model species representing the group of small mammals, differing from one another not only in terms of important elements of ecology, but also as to their history and manner of settling the city. However, irrespectively of those differences, our analysis of microsatellite polymorphism within and among urban and non-urban populations of the two mouse species suggests that the urban environment increases genetic diversity among local populations living in suitable habitat patches within a city, and between those inhabiting city and rural areas. It seems that suitably planned areas at the city borders could play an important role as ‘gateways’ through which individuals from suburban populations could migrate into the city and join urban populations. This would permit conservation of high diversity of species in cities, and would prevent disadvantageous decreases in genetic variability in populations of particular species due to the fragmentation and isolation of local urban local populations (Saarikivi et al. 2013). However, it is necessary to carefully plan the urban infrastructure so as to preserve a network of ecological corridors connecting local green areas (Szacki et al. 1994). Our data suggest that the genetic drift and founders’ effects are the main processes influencing the genetic structure of urban populations. For this reason, it seems to be crucial for the conservation of genetic diversity to enable even small groups of individuals to migrate between particular subpopulations within the city. This is a strong argument in favour of the construction of ecological networks in cities in order to prevent the loss of genetic diversity in synurbic species populations and to improve the quality of life of people inhabiting these cities.
References
Aars J, Ims RA, Liu H-P, Mulvey M, Smith MH (1998) Bank voles in linear habitats show restricted gene flow as revealed by mitochondrial DNA (mtDNA). Mol Ecol 7:1383–1389
Andrzejewski R, Babinska-Werka J, Gliwicz J, Goszczynski J (1978) Synurbization processes in population of A. agrarius. I. Characteristics of populations in urbanization gradient. Acta Theriol 23:341–358
Avise JC (1994) Molecular Markers, Natural History and Evolution. Chapman & Hall, New York
Babińska-Werka J, Malinowska B (2008) Synurbization of the yellow-necked mouse A. flavicollis in Warsaw. In: Indykiewicz P, Jerzak L, Barczak T (eds) Fauna of cities. Preservation of biodiversity in cities. ATR, Bydgoszcz, pp. 144–150 (in Polish with English abstract)
Baker RJ, Makova KD, Chesser RK (1999) Microsatellites indicate a high frequency of multiple paternity in Apodemus (Rodentia). Mol Ecol 8(1):107–111
Bjӧrklund M, Ruiz I, Senar J (2010) Genetic differentiation in the urban habitat: the great tits (Parus major) of the parks of Barcelona city. Biol J Linn Soc 99:9–19
Booth W, Montgomery WI, Prodöhl PA (2009) Spatial genetic structuring in a vagile species, the European wood mouse. J Zool 279:219–228
Bujalska G (1981) Reproduction strategies in populations of Microtus arvalis (Pall.) and Apodemus agrarius (Pall.) inhabiting farmland. Pol Ecol Stud 7:229–243
Bujalska G, Grüm L (2005) Reproduction strategy in an island population of yellow-necked mice. Popul Ecol 47:151–154. doi:10.1007/s10144-005-0219-y
Bujalska G, Grüm L (2006) Winter survival of A. flavicollis in Crabapple Island (NE Poland). Hystrix Italian J Mammal (n.s.) 17(2):173–177
Bujalska G, Grüm L (2008) Interaction between populations of the bank vole and the yellow-necked mouse. Ann Zool Fenn 45:248–254
Chakraborty R, Nei M (1977) Bottleneck effects on average heterozygosity and genetic distance with the stepwise-mutation model. Evolution 31:347–356
Chiappero MB, Panzetta-Dutari GM, Gomez D, Castillo E, Polop JJ, Gardenal CN (2011) Contrasting genetic structure of urban and rural populations of the wild rodent Calomys musculinus (Cricetidae, Sigmodontinae). Mamm Biol 76:41–50
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Dearborn DC, Kark S (2010) Motivations for conserving urban biodiversity. Conserv Biol 24(2):432–440. doi:10.1111/j.1523-1739.2009.01328.x
Delaney KS, Riley SPD, Fisher RN (2010) A rapid, strong and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS One 5(9):e12767
Desender K, Small E, Gaublomme E, Verdyck P (2005) Rural-urban gradients and the population genetic structure of woodland ground beetles. Conserv Genet 6:51–62
Di Rienzo A, Peterson CA, Garza JC, Valdes AM, Slatkin M, Freimer NB (1994) Mutational processes of simple-sequence repeat loci in human populations. P Natl Acad Sci USA 91:3166–3170. doi:10.1073/pnas.91.8.3166
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Earl DA, von Holdt BM (2011) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Gen Res 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Gardner-Santana LC, Norris DE, Fornadel CM, Hinson ER, Klein SL, Glass GE (2009) Commensal ecology, urban landscapes, and their influence on the genetic characteristics of city-dwelling Norway rats (Rattus norvegicus). Mol Ecol 18:2766–2778
Gerlach G, Musolf K (2000) Fragmentation of landscape as a cause for genetic subdivision in bank voles. Conserv Biol 14:1066–1074
Gliwicz J (1980) Ecological aspects of synurbization of Striped Field Mouse A. agrarius (Pall.). Wiad Ekol 26:185–196
Gockel J, Harr B, Schlötterer C, Arnold W, Gerlach G, Tautz D (1997) Isolation and characterization of microsatellite loci from A. flavicollis (Rodentia, Muridae) and Myodes glareolus (Rodentia, Cricetidae). Mol Ecol 6:597–599
Goddard MA, Dougill AJ, Benton TG (2010) Scaling up from gardens: biodiversity conservation in urban environments. Trends Ecol Evol 25:90–98
Gortat T, Rutkowski R, Gryczyńska-Siemiątkowska A, Kozakiewicz A, Kozakiewicz M (2013) Genetic structure in urban and rural populations of A. agrarius in Poland. Mamm Biol 78:171–177
Gortat T, Barkowska M, Gryczyńska-Siemiątkowska A, Pieniążek A, Kozakiewicz A, Kozakiewicz M (2014) The effects of urbanization – small mammal communities in a gradient of human pressure in Warsaw city, Poland. Pol J Ecol 62:163–172
Gortat T, Rutkowski R, Gryczyńska-Siemiątkowska A, Kozakiewicz A, Pieniążek A, Kozakiewicz M (2015a) Anthropopressure gradients and the population genetic structure of A. agrarius. Conserv Genet 16:649–659
Gortat T, Rutkowski R, Gryczyńska-Siemiątkowska A, Kozakiewicz A, Kozakiewicz M (2015b) Genetic variability in island populations of two rodent species: bank vole (Myodes glareolus) and yellow-necked mouse (Apodemus favicollis). Ann Zool Fenn 52:145–159
Goudet J (2001) FSTAT V2.9.3, a program to estimate and test gene diversities and fixation indices. http://www.unil.ch/izea/softwares/fstat.htlm
Gryczyńska-Siemiątkowska A, Gortat T, Rutkowski R, Pomorski J, Kozakiewicz A, Kozakiewicz M (2008) Multiple paternity in a wild population of yellow-necked mouse Apodemus flavicollis. Acta Theriol 53(3):251–258
Harr B, Musolf K, Gerlach G (2000) Characterization and isolation of DNA microsatellite primers in wood mice (Apodemus sylvaticus, Rodentia). Mol Ecol 9:1664–1665
Hitchings SP, Beebee TJC (1998) Loss of genetic diversity and fitness in Common Toad (Bufo bufo) populations isolated by inimical habitat. J Evol Biol 11:269–283
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. doi:10.1093/bioinformatics/btn129
Jombart T, Devillard S, Dufour A-B, Pontier D (2008) Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101:92–103. doi:10.1038/hdy.2008.34
Kadlec T, Benes J, Jarosik V, Konvicka M (2008) Revisiting urban refuges: Changes of butterfly and burnet fauna in Prague reserves over three decades. Landsc Urban Plan 85:1–11
Kajdacsi B, Costa F, Hyseni C, Porter F, Brown J, Gorete R, Farias H, Reis MG, Childs JE, Ko AI, Caccone A (2013) Urban population genetics of slum-dwelling rats (Rattus norvegicus) in Salvador, Brazil. Mol Ecol 22(20):5056–5070
Kalinowski ST (2005) A computer program for performing rarefaction on measures of alleli diversity. Mol Ecol Notes 5:187–189
Lidicker WZ Jr (1975) The role of dispersal in the demography of small mammals. In: Petrusewicz K, Golley FB, Ryszkowski L (eds) Small Mammals: productivity and dynamics of populations. Cambridge University Press, Cambridge, pp. 103–128
Makova KD, Patton JC, Krysanov EYU, Chesser RK, Baker RJ (1998) Microsatellite markers in wood mouse and striped field mouse (genus Apodemus). Mol Ecol 7:247–249
McCauley DE (1991) Genetic consequences of local population extinction and recolonization. Trends Ecol Evol 6:5–8
Mikulič P, Pišút P (2012) Genetic structure of the marsh frog (Phelophylax ridibundus) populations in urban landscape. Eur J Wildl Res 58:833–845
Munshi-South J, Kharchenko K (2010) Rapid, pervasive genetic differentiation of urban white-footed mouse (Peromyscus leucopus) populations in New York City. Mol Ecol 19:4242–4254
Munshi-South J, Nagy C (2014) Urban park characteristics, genetic variation, and historical demography of white-footed mouse (Peromyscus leucopus) populations in New York City. Peer J 2:e310. doi:10.7717/peerj.310
Munshi-South J, Zak Y, Pehek E (2013) Conservation genetics of extremely isolated urban populations of the Northern Dusky Salamander (Desmognathus fuscus) in New York City. Peer J 1:e64. doi:10.7717/peerj.64
Muratet A, Machon N, Jiguet F, Moret J, Porcher E (2007) The role of urban structures in the distribution of wasteland flora in the Greater Paris Area, France. Ecosystems 10(4):661–671
Nei M, Roychoudhury AK (1974) Sampling variances of heterozygosity and genetic distance. Genetics 76:379–390
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Noël S, Lapointe F-J (2010) Urban conservation genetics: study of a terrestrial salamander in the city. Biol Conserv 143:2823–2831
Noël S, Ouellet M, Galois P, Lapontaine F-J (2007) Impact of urban fragmentation on the genetic structure of the eastern red-backed salamander. Conserv Genet 8:599–606
Ohnishi N, Ishibashi Y, Saitoh T, Abe S, Yoshida MC (1998) Polymorphic microsatellite DNA markers in the Japanese wood mouse Apodemus argenteus. Mol Ecol 7:1431–1432
Paekall R, Smouse PE (2001) GenAlEx V5: Genetic Analysis in Excel. Population genetic software for teaching and research. http://www.anu.ed.au/BoZo/GenAlEx/
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pucek Z (1984) Keys to vertebrates of Poland: Mammals. PWN, Poland, p. 357
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 43:258–275
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Her 86:248–249
Ricketts T, Imhoff M (2003) Biodiversity, urban areas, and agriculture: locating priority ecoregions for conservation. Conserv Ecol 8(2):1 (online) URL: http://www.consecol.org/vol8/iss2/art1/
Rosenberg NA (2004) DISTRUCT, a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (1997) Genetic differentiation and estimation of gene flow from F- statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2008) Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Rubin CS, Warner RE, Bouzat JL, Paige KN (2001) Population genetic structure of Blanding's turtles (Emydoidea blandingii) in an urban landscape. Biol Conserv 99:323–330
Rutkowski R, Rejt Ł, Tereba A, Gryczyńska-Siemiątkowska A, Janic B (2009) Population genetic structure of the European kestrel Falco tinnunculus in Central Poland. Eur J Wildl Res 56:297–305
Saarikivi J, Knopp T, Granroth A, Merilä J (2013) The role of golf courses in maintaining genetic connectivity between common frog (Rana temporaria) populations in an urban setting. Conserv Genet 14:1057. doi:10.1007/s10592-013-0495-6
Slatkin M (1977) Gene flow and genetic drift in a species subject to frequent local extinctions. Theor Popul Biol 12:253–262
Sudnik-Wójcikowska B (1988) Flora synanthropization and anthropopressure zones in a large urban agglomeration (exemplified by Warsaw). Flora 180:259–265
Suminski SM (1922) Fauna of Warsaw. Ziemia 12:328–335 (in Polish)
Szacki J, Głowacka I, Liro A, Matuszkiewicz A (1994) The role of connectivity in the urban landscape: some results of research. Memorabilia Zool 49:49–56
Unfried TM, Huuser L, Marzluff JM (2013) Effects of urbanization on Song Sparrow (Melospiza melodia) population connectivity. Conserv Genet 14(1):41–53. doi:10.1007/s10592-012-0422-2
United Nations (UNDP) (2011). Human Development Report. Sustainability and Equity: A Better Future for All.1 UN Plaza, New York, NY 10017, USA. pp 179. pdf. http://hdr.undp.org/sites/default/files/reports/271/hdr_2011_en_complete
Vangestel C, Mergeay J, Dawson DA, Vandomme V, Lens L (2011) Spatial heterogenity in genetic relatedness among house sparrows along an urban-rural gradient as revealed by individual-based analysis. Mol Ecol 20:4643–4653
VIEP (2013) The State of the Environment in Mazowieckie Voivodship in 2012. Voivodship Inspectorate of Environmental Protection, Warsaw, p. 173 (in Polish)
Walecki A (1881) Mammalian fauna of Warsaw in relation to fauna of the whole country. Pamiętnik Fizjograficzny 1:268–291 (in Polish)
Wandeler P, Funk M, Largiader R, Gloor S, Breitenmoser U (2003) The city-fox phenomenon: genetic consequences of a recent colonization of urban habitat. Mol Ecol 12:647–656
Waples RS, Do C (2008) ldne: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whitlock MC, McCauley DE (1990) Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44:1717–1742
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wood BC, Pullin AS (2002) Persistence of species in a fragmented urban landscape: the importance of dispersal ability and habitat availability for grassland butterflies. Biodivers Conserv 11:1451–1468
Zak Y, Pehek E (2013) Conservation genetics of extremely isolated urban populations of the northern dusky salamander (Desmognathus fuscus) in New York City. Peer J 1:e64. doi:10.7717/peerj.64
Acknowledgements
The study was carried out at the Biological and Chemical Research Centre, University of Warsaw, established within the project co-financed by European Union from the European Regional Development Fund under the Operational Programme Innovative Economy, 2007 – 2013. The study was financed by the National Science Centre of Poland (Grant N N304 169539).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 15 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gortat, T., Rutkowski, R., Gryczynska, A. et al. The spatial genetic structure of the yellow-necked mouse in an urban environment – a recent invader vs. a closely related permanent inhabitant. Urban Ecosyst 20, 581–594 (2017). https://doi.org/10.1007/s11252-016-0620-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11252-016-0620-7