Abstract
Urban development can fragment and degrade habitat, and such habitat alterations can have profound effects on wildlife, including influencing population genetics. We used nine microsatellite loci to determine the effects of urbanization on genetic diversity and genetic structure in a native small mammal, Merriam’s kangaroo rat, in areas in and around Las Cruces, NM, an expanding low density urban center. We found that Merriam’s kangaroo rats in urban areas had increased genetic differentiation among populations as compared to wildland animals, and detected some evidence of lower genetic diversity in urban areas, indicating that the population genetics of a common and abundant wildlife species can be impacted by low density urbanization. Our results suggest that although abundant and common wildlife such as Merriam’s kangaroo rats may persist in urban environments, these animals may still be influenced by more subtle effects of urbanization, such as genetic isolation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Urban development fragments habitats via roads, houses, and buildings; such fragmentation can increase isolation of remnant habitats and wildlife populations. Isolated wildlife populations inhabiting habitat fragments may experience a decrease in genetic diversity within populations, and an increase in genetic differentiation among populations (Keyghobadi 2007). Genetic diversity is important for future population viability, particularly in the face of environmental change (Reed and Frankham 2003; Frankham 2005; Lourenco et al. 2017). In addition, small populations may experience magnified effects of genetic drift and reduced gene flow (Segelbacher et al. 2003; Reed and Hobbs 2004; Frankham 2005; Keely et al. 2015). In extreme cases, genetic drift, reduced gene flow, and small population size can lead to inbreeding depression (Keller and Waller 2002; Reed and Frankham 2003; Frankham 2005; Lourenco et al. 2017).
Urbanization has been shown to affect genetic structure in a variety of species, including mesocarnivores, small mammals, birds, non-avian reptiles, and amphibians (Riley et al. 2006; Delaney et al. 2010; Munshi-South and Kharchenko 2010; Noël and Lapointe 2010; Santonastaso et al. 2012; Munshi-South et al. 2013). Specifically, high levels of urban development have been correlated with decreased genetic diversity, increased genetic structure, and decreased gene flow in terrestrial animals (Riley et al. 2006; Magle et al. 2010; Munshi-South and Kharchenko 2010; Munshi-South et al. 2013). For example, populations of Stephen’s kangaroo rats (Dipodomys stephensi) in southern California, white-footed mice (Peromyscus leucopus) in New York City, and black-tailed prairie dogs (Cynomys ludovicianus) in Denver, Colorado all exhibited increased population genetic structure in urban areas as compared to populations in wildland areas (McClenaghan and Truesdale 2002; Magle et al. 2010; Munshi-South et al. 2016). In addition, white-footed mouse populations demonstrated decreased genetic diversity (Munshi-South et al. 2016), and black-tailed prairie dog colonies demonstrated reduced movement rates between urban colonies (Magle et al. 2010). Most of these studies have been conducted in highly urbanized areas, consistent with trends across the field of urban ecology; a recent review found that 85% of urban ecology research was conducted in high density urban areas, such as Phoenix, AZ (Magle et al. 2012). Studies of genetic structure in suburban, low density or newly urbanized sites are less common. However, in the few studies that have been conducted in less-intensely urbanized areas, genetic structure has been detected: suburban Japanese field mice (Apodemus speciosus) populations were found to have significant genetic differentiation as compared to wildland populations (Hirota et al. 2004), and across an urbanization gradient, striped field mice (Apodemus agrarius) in the most urbanized areas demonstrated significant genetic structuring as compared to suburban populations (Gortat et al. 2015).
Despite the paucity of studies in low density urban areas, the importance of such areas for landscape conservation and management has been recognized (Theobald 2001). These landscapes are thought to be important for wildlife, both as habitat and as stepping stones between undeveloped lands and highly urbanized environments (Magle et al. 2012). With the continuous low density urban expansion characteristic of the southwestern and southeastern United States (Theobald 2005), it is imperative to understand how animals are influenced by such urban expansion. Of equal importance is the evaluation of the potential impact of such low density urbanization on wildlife population genetics. We investigated the effects of rapidly expanding low density urbanization on genetic diversity, genetic structure and gene flow in Merriam’s kangaroo rats (Dipodomys merriami). We predicted that kangaroo rat populations in low density urban sites would have lower genetic diversity, increased genetic structure, and less gene flow among sites than animals living in wildland areas.
Methods
Study species
Kangaroo rats (genus Dipodomys) are common throughout the deserts and grasslands of the western and southwestern United States (Kennedy and Schnell 1978; Sullivan and Best 1997). We focused on Merriam’s kangaroo rat (D. merriami), a granivorous Heteromyid rodent that is abundant and found in desert scrub habitats in both wildland and urban environments across the southwestern United States (Germaine et al. 2001; DaVanon et al. 2016; Hurtado and Mabry 2017). However, to date relatively few studies have capitalized on D. merriami as a study organism for evaluating the impacts of rapidly growing low density urban development on wildlife population genetics.
Study area
This study was conducted in and around the City of Las Cruces, New Mexico, USA (32° 19′ 35.7414″, − 106° 46′ 31.569″). Las Cruces is a small city that has grown rapidly in recent decades, with a human population that increased by more than 35% from 2000 to 2014 (from 74,267 to 101,408; U.S. Census Bureau 2015). The city encompasses several urban parks and open spaces that retain natural vegetation but are surrounded by paved roads, homes, and businesses. The city is surrounded by undeveloped desert areas, which are primarily managed by the U.S. Bureau of Land Management.
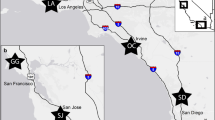
We established 10 study sites in both urban (n = 5 within Las Cruces city limits) and wildland environments (n = 5 on federal and state properties surrounding Las Cruces; Fig. 1). All sites were large enough to accommodate a 100 m by 100 m live-trapping grid, contained similar native vegetation, and similarly composed soils (Hurtado 2017; Hurtado and Mabry 2017). Wildland sites were located at least 500 m from paved roads, and all study sites were at least 1 km from other sites (Hurtado and Mabry 2017). We quantified the proportion impervious surface (i.e. roads, cement, and buildings) within 500 m of the center of each site using Landsat Thematic Mapper (LTM) imagery and a modified Normalized Difference Vegetation Index (NDVI) to create an urbanization index which we used as a proxy for urbanization (NASA Earth Observatory 2016; Yuan and Bauer 2007; Song et al. 2016). A strong correlation between the number of buildings at a site and the urbanization index indicated that this index was a good measure of urbanization (Hurtado and Mabry 2017). All processing was conducted in ArcGIS 10.1 (ESRI, Environmental Systems Research Institute 2012 Redlands, California). We selected study sites that were as similar as possible in all aspects other than the level of surrounding human development and proportion of impervious surface. There were no differences in vegetative cover (percent cover by shrubs, forbs, grass, litter, bare ground, or rock) between urban and wildland sites, but the urbanization index for urban sites was approximately an order of magnitude larger (mean ± 1 SE = 0.33 ± 0.01) than that of wildland sites (0.03 ± 0.01; Hurtado 2017).
Map of Las Cruces, NM, showing roads and placement of study sites. Circles are the 500 m buffers around trapping sites that were used to calculate the urbanization index for each site. Cross-hatched buffers are urban sites and non-shaded buffers are wildland sites. With the exception of site AH, all sites are located on the same side of major highways: east of Interstates 10 and 25, and south of NM Highway 70. Site AH is located near the intersection of Interstates 10 and 25, and is separated from all other sites by a total of 8 lanes of traffic: 4 for I-25 and 2 each for parallel surface streets
Live trapping
Merriam’s kangaroo rats were live trapped over three years, May to November 2013, June to September 2014, and May to October 2015. Trapping methods follow those reported in Hurtado and Mabry (2017). In 2013, traps were set in a 10 × 10 grid with 10 m spacing between traps. In 2014 and 2015, traps were set within the bounds of the 1 ha trapping grids used in 2013, but traps were placed along kangaroo rat trails and outside burrows so as to maximize capture success. Due to possible reduced trappability on moonlit nights, Merriam’s kangaroo rats were trapped only during the 6 days before and after the new moon. Sherman live traps (model XLKGDT with short doors to avoid catching the long tails of Merriam’s kangaroo rats; H.B. Sherman Co., Tallahassee, FL) were used in this study. Traps were baited with a millet–sunflower seed mixture and placed under vegetation before sunset, to protect animals from direct sunlight. Traps were checked between 01:00 and 08:00 h; traps were checked earlier in the morning at sites that had high human visitation, or on cold nights (projected to be below 4.5 °C). In urban areas, traps were removed daily, replaced, and reset before sunset. In wildlands, traps were checked, closed, and left in place during the day. Each captured Merriam’s kangaroo rat was uniquely marked with numbered Monel ear tags (National Band and Tag, Newport, KY), and standard data were recorded (e.g. sex, reproductive condition, foot length, mass), and tissue samples collected (ear snip) before release at the site of capture. Tissue samples were placed in individually labeled 2 mL tubes that were put on ice while in the field and then transferred to a − 20 °C freezer in the laboratory until processing. All research procedures were consistent with the guidelines of the American Society of Mammalogists (Sikes et al. 2011) and conducted under an approved New Mexico State University IACUC protocol (protocol 2013-014); no permits were required from the New Mexico Department of Game and Fish.
DNA extraction and microsatellite development
Total genomic DNA was extracted using the DNeasy Blood and Tissue kit (QIAGEN, Hilden, Germany), following the manufacturer’s protocol. We quantified DNA concentration using a Nanodrop spectrophotometer. Previously published microsatellites that were developed for other Dipodomys species (Vega et al. 2007; Meshriy et al. 2011) failed to amplify well for Merriam’s kangaroo rat (L.K. Howard, unpublished data). Therefore, species-specific microsatellites for Merriam’s kangaroo rat were developed by the University of Georgia Savannah River Ecological Laboratory (SREL) genetics laboratory. We then screened 48 of the newly-identified microsatellites for amplification and variability in our lab, using DNA from 16 Merriam’s kangaroo rats. We then selected the most highly-variable loci, all of which had repeat units of 4–6 nucleotides, and ordered fluorescently-labeled primers to conduct genotyping of all samples.
Microsatellite amplification
For each sample, we amplified 11 microsatellite loci in individual 12.5 µL PCR reactions containing 0.5 µ of each primer (with a fluorescent label on the 5′ end of each forward primer), 10 mM 10× AmpliTaq Gold buffer, 0.15 mM deoxynuclecoside triphosphates (Invitrogen), 0.1 units of Taq (AmpliTaq Gold, Applied Biosystems), 25.0 µg/mL bovine serum albumin, a locus-specific amount of MgCl2 (Table 1), nuclease-free water, and 1 µL of template DNA (mean concentration ± 1 SE: 24.88 ± 2.98 ng/µL). PCRs were conducted on a BIO-RAD C1000 Thermal Cycler (Hercules, California) with an initial denaturation at 94.0 °C for 540 s, followed by 40 cycles of 94.0 °C for 30 s, a locus-specific annealing temperature for 30 s (decrementing by 0.1 °C each cycle), 72 °C for 30 s, then 30 cycles of 94.0° for 30 s, six degrees below the locus-specific annealing temperature for 30 s, 72.0° for 30 s, and a final extension step of 72.0 °C for 420 s. PCR products were combined into 3 sets with LIZ 500 size standard (Applied Biosystems), and fragment analysis was conducted on an Applied Biosystems 3130xL Genetic Analyzer. We used GeneMapper v. 4 (Thermo Fisher Scientific, Waltham, MA) to score alleles and Flexibin to bin alleles into discrete size categories (Amos et al. 2007).
Analysis of genetic diversity and population genetic structure
We used GenAlEx (ver. 6.503, Peakall and Smouse 2012) to characterize allelic diversity at each locus, including the number of unique alleles and observed and expected heterozygosity across study sites (Table 1). We also determined the number of alleles, private alleles, effective alleles, and observed and expected heterozygosity across all loci for each site, and compared the values for all of these variables between urban and wildland sites using non-parametric Wilcoxon rank sums tests.
To evaluate population genetic structure, we initially used GenAlEx 6.5 (Peakall and Smouse 2012) to calculate pairwise Fst and Nei’s G’st, and to determine P-values for each pair-wise comparison, using 9999 permutations. We then applied a false discovery rate (FDR; Benjamini and Hochberg 1995) of 0.1 to assess the statistical significance of the resulting 45 pair-wise comparisons. We used the program Structure (which uses Bayesian assignment to infer population clusters; Hubisz et al. 2009; Pritchard et al. 2000), and Structure Harvester to determine the most likely number of genetic clusters (K), using the Evanno method (Evanno et al. 2005). Initial analyses did not indicate a strong effect of variation in sample size among sites on the number of clusters identified by Structure; analyses that included either all 212 genotyped animals (with 9–35 samples/site), or a reduced data set from which individuals were randomly removed from more thoroughly sampled sites until n = 19 (yielding 9–19 samples/site and a total n = 169) yielded similar results, and we present results from the analysis of the full data set. We conducted Structure analyses using values of K from 1 to 15, the admixed ancestry model, correlated allele frequencies, a burnin of 100,000 followed by 1,000,000 MCMC iterations, with 10 runs for each value of K. To refine estimates of cluster membership for each sample, we then conducted a longer Structure run for the single most-likely value of K, with all parameters as above, but increasing the number of MCMC iterations to 5,000,000. We also used the program Geneland (Guillot et al. 2005; Guillot and Santos 2009), which incorporates spatial information into a Bayesian clustering algorithm to determine the number of genetic clusters present. As in the Structure analysis, we found that both the full dataset and a reduced dataset with more even sampling across sites yielded similar results, and we present results from the full dataset. In Geneland, we used the spatial model with correlated allele frequencies, location uncertainty of 25 m, K of 1 to 15, 1,000,000 MCMC iterations, thinning every 1000 iterations, with 10 runs per value of K. We then post-processed with a secondary burn-in of 500. After the first set of Geneland runs to identify the most likely number of genetic clusters, we conducted another model run which used the same parameters as above, but only for the most likely value of K, and increasing the number of MCMC repetitions to 5,000,000. This final Geneland run refined our estimates of proportional membership of each individual to each identified genetic cluster.
Assignment tests
To indirectly evaluate potential migration (gene flow) between sites, and to further evaluate population structuring between sites, we conducted an assignment test in GenAlEx, using the leave one out option. All ten populations and all 212 individuals were used in the assignment test.
Isolation by distance
We used a Mantel test to evaluate the relationship between geographic distance and genetic distance among populations. We used GenAlEx to create matrices for genetic and geographic distance, with one set of matrices that included all populations, and separate matrices for urban and wildland populations; we then ran Mantel tests with 999 permutations in R 3.1.2 (R Development Core Team 2016), using the package ada4 version 1.7-6 (Chessel et al. 2002).
Results
Genetic diversity
Two microsatellite loci were dropped from analysis due to both linkage disequilibrium and non-conformance to Hardy–Weinberg equilibrium (DIME 30 and DIME 10); both loci were linked to DIME 4. The remaining 9 polymorphic microsatellite loci were used in all analyses. The overall mean number of alleles per locus was 14.22 ± 0.85 (mean ± 1 SE, range 10–18). The mean number of alleles per locus in urban sites was 7.44 ± 0.18, which was slightly lower than the value for wildland sites: 8.69 ± 0.55 (Table 2; Wilcoxon rank sums test, W = 3.5, P = 0.07). Private alleles were detected in all 5 urban sites, and in 3 out of 5 wildland sites (Table 2). No statistical difference was found in the number of private alleles between urban and wildland sites (Wilcoxon rank sums test, W = 12.5, P = 1.00). The mean number of effective alleles per site was higher in wildland sites (5.71 ± 0.17) than urban sites (4.52 ± 0.11; Wilcoxon rank sums test, W = 0, P = 0.008). Observed heterozygosity was not different between urban and wildland sites (Wilcoxon rank sums test, W = 13.5, P = 0.92), but expected heterozygosity was higher in wildland sites (Wilcoxon rank sums test, W = 1.5, P = 0.03).
Population structure
Results of Fst and G’st analyses were similar; we report Fst results here. Pair-wise Fst indicated that each urban site was genetically dissimilar from all other sites (both urban and wildland), while wildland sites did not generally appear to be significantly differentiated from each other, with the exception of site W.LA (Table 3). The Δ(K) method implemented in Structure Harvester analysis indicated that the most likely value of K was 4 (Fig. 2). However, there was no clear evidence of distinct population clusters (Fig. 3); most individuals were assigned some proportional membership to all 4 clusters, suggesting a lack of strong genetic structure.
In contrast to the Structure analysis, Geneland detected evidence of genetic structure, inferring eight clusters (Fig. 4). There were 6 clusters that were predominantly associated with a single sampling site; one for each of 5 urban sites, and for a single wildland site, W.LA (Fig. 4). The 2 remaining genetic clusters (2, 5) represented the other 4 wildland sites (Fig. 4). These results are consistent with those from the Fst analysis.
Maps showing eight genetic clusters identified by Geneland. Black dots correspond to the study sites shown on the map in Fig. 1. Lighter shading (yellow) is associated with increased mean proportional membership of kangaroo rats sampled at a site to a genetic cluster, and darker shading (red) is associated with lower proportional membership to a cluster; isoclines show areas of increasing proportional membership to a cluster. Note that shading and isoclines are not always directly comparable among clusters. Clusters 1, 3, 4, 6–8 are each associated with a single sampling site, while animals sampled at 4 of the 5 wildland sites are associated with genetic clusters 2 and 5. (A) Cluster 1: U.DACC, (B) Cluster 2: W.AG, W.GEO, W. SS, W. TT, (C) Cluster 3: U.CD, (D) U.AH, (E) Cluster 5: W.AG, W.GEO, W. SS, W. TT, (F) Cluster 6: W.LA, (G) Cluster 7: U.DT, (H) Cluster 8, U.PP. (Color figure online)
A Mantel test examining all sites (urban and wildland) indicated that there was no correlation between geographic distance and genetic distance among populations (r = 0.263, P = 0.47). Similarly, separate Mantel tests for urban and wildland populations did not detect a statistically significant relationship that would be consistent with isolation-by-distance (urban: r = 0.07, P = 0.26; wildland: r = − 0.584, P = 0.83).
Population assignment
Success at assigning individuals to their population of capture ranged from 0 to 90.3% (Table 4). For urban sites, individuals were assigned to their population of capture on average in 72% of cases (range 57.2 to 90.3%). In contrast, in wildland sites, individuals were assigned to their population of capture much less frequently: on average, 27.7% of the time (range 0 to 54.3%). Urban sites had a high correct assignment rate and few animals assigned to other populations. Wildland sites had a low correct assignment rate and many animals were assigned to other populations.
Discussion
We found strong evidence of genetic structure among Merriam’s kangaroo rats living in urban sites, although the differences in genetic diversity between urban and wildland populations were somewhat less clear. Results indicated that there was reduced gene flow among urban sites, these results support the hypothesis that rapidly-growing low density urbanization has influenced genetic structure of Merriam’s kangaroo rat over a very short time scale. Our results are especially striking given that the average age of urban development near our urban study sites was just over 20 years, indicating that relatively new, low density urbanization may affect population genetics and gene flow, even in species that are abundant in urban areas. Consideration of the effects of low density urban development on the population genetics of wild animals is of particular importance if management and conservation strategies view these low density areas as potential wildlife habitat in higher urban density areas, or as stepping stones from adjacent wildlands into high density urban areas (Theobald 2001; Magle et al. 2012).
We used multiple methods to assess genetic structure in Merriam’s kangaroo rats, including a straightforward comparison of pair-wise Fst among sampling sites, and Bayesian clustering algorithms implemented in Structure and Geneland. These methods yielded differing results: Structure detected genetic clustering, but the clusters did not align with our sampling sites. In contrast, both Geneland and Fst comparisons indicated that kangaroo rats from all 5 urban sites and 1 of the wildland sites were genetically distinct from those sampled at the other 4 wildland sites. The inclusion of spatial information in Geneland yields increased power to detect low-level genetic structuring (Guillot 2008; François and Durand 2010); interestingly, Fst analysis also inherently incorporates spatial information (because populations are pre-defined), although it does not include the relative spatial positions and distances between sampling sites. On average, our wildland sampling sites were not closer together than our urban sites (Fig. 1), and some wildland sites were much closer to urban sites than they were to other wildland sites (e.g., W.SS is much closer to U.DACC and U.DT than any other sites, yet is not genetically similar; Fig. 1). Along with the lack of evidence of isolation-by-distance, these results suggest that urbanization rather than distance per se is responsible for the patterns of genetic structure that we observed.
However, despite the detection of genetic structure, we did not detect substantial differences in genetic diversity between urban and wildland sampling sites. Wildland sites had a higher number of effective alleles, higher expected heterozygosity, and a slightly higher number of alleles/locus, but the number of private alleles and observed heterozygosity were similar between urbanization classes (Table 2). Taken together, these results suggest that in the relatively short time that our low-density urban sites have been developed (~ 20 years), gene flow among urban populations has been reduced, but substantial genetic diversity has not yet been lost. However, lower numbers of effective alleles per locus in urban sites may suggest that the expected loss of genetic diversity with increasing urbanization has begun. Re-sampling kangaroo rat genetic diversity at this sites 10–20 years in the future would provide a test of this prediction, assuming that sampling sites remain intact; site U.AH was developed shortly after field collections for this study were complete.
In summary, our results show that the population genetic structure of a common native rodent has been affected by rapidly growing low density urban areas, the most rapidly growing type of urban development in the nation (Theobald 2005). Results such as ours potentially indicate that other common species may also be impacted by low density urban development across the landscape. Further, rarer (i.e. threatened and endangered) or more isolated wildlife may face more issues in urbanizing environments due to genetic drift and reduced gene flow (Reed and Hobbs 2004; Keely et al. 2015). This should be of concern to managers and conservation planners. More research on the impacts of low density urban areas on wildlife, and on the development and management of urban green spaces (with native vegetation) specifically designed for wildlife connectivity and gene flow is needed.
References
Amos W, Hoffman JI, Frodsham A, Zhang L, Best S, Hill AVS (2007) Automated binning of microsatellite alleles: problems and solutions. Mol Ecol Notes 7:10–14
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Chessel D, Dufour A-B, Dray S, Lobry JR, Ollier S, Pavoine S, Thioulouse J (2002) ada4: analysis of ecological data: exploratory and euclidean methods in environmental sciences. R package version 1.7-6. http://pbil.univ-lyon1.fr/ADE-4/. Accessed Mar 2017
DaVanon KA, Howard LK, Mabry KE, Schooley RL, Bestelmeyer BT (2016) Effects of exurban development on trophic interactions in a desert landscape. Landsc Ecol 31:2343–2354
Delaney KS, Riley SPD, Fisher RN (2010) A rapid, strong, and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS ONE 5:1–11
ESRI, Environmental Systems Research Institute (2012) Redlands, California. https://www.esri.com/en-us/home. Accessed June 2012
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
François O, Durand E (2010) Spatially explicit Bayesian clustering models in population genetics. Mol Ecol Res 10:773–784
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Germaine SS, Schweinsburg RE, Germaine HL (2001) Effects of residential density on Sonoran Desert nocturnal rodents. Urban Ecosyst 5:179–185
Gortat T, Rutkowski R, Gryczyńska A, Pieniążek A, Kozakiewicz A, Kozakiewicz M (2015) Anthropopressure gradients and the population genetic structure of Apodemus agrarius. Conserv Gen 16:649–659
Guillot G (2008) Inference of structure in subdivided populations at low levels of genetic differentiation—the correlated allele frequencies model revisited. Bioinformatics 24:2222–2228
Guillot G, Santos F (2009) A computer program to simulate multilocus genotype data with spatially auto-correlated allele frequencies. Mol Ecol Res 9:1112–1120
Guillot G, Mortier F, Estoup A (2005) Geneland: a program for landscape genetics. Mol Ecol Notes 5:712–715
Hirota T, Hirohata T, Mashima H, Satoh T, Obara Y (2004) Population structure of the large Japanese field mouse, Apodemus speciosus (Rodentia: Muridae), in suburban landscape, based on mitochondrial D-loop sequences. Mol Ecol 13:3275–3282
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Res 9:1322–1332
Hurtado G (2017) Effects of urbanization on behavior, parasite prevalence, and population genetics of a southwestern mammal. PhD dissertation, New Mexico State University, Las Cruces, NM, USA
Hurtado G, Mabry KE (2017) Aggression and boldness in Merriam’s kangaroo rat: an urban-tolerant species? J Mammal 98:410–418
Keely CC, Hale JM, Heard GW, Pasrris KM, Summer J, Hamer AJ, Melville J (2015) Genetic structure and diversity of the endangered growling grass frog in a rapidly urbanizing region. R Soc Open Sci 2:140255
Keller L, Waller D (2002) Interbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Kennedy ML, Schnell GD (1978) Geographic variation and sexual dimorphism in Ord’s kangaroo rat, Dipodomys ordii. J Mammal 59:45–59
Keyghobadi N (2007) The genetic implications of habitat fragmentation for animals. Can J Zool 85:1049–1064
Lourenco A, Alvarez D, Wang IJ, Velo-Anton G (2017) Trapped within the city: integrating demography, time since isolation and population-specific traits to assess the genetic effects of urbanization. Mol Ecol 26:1498–1514
Magle SB, Ruell E, Antolin MF, Crooks KR (2010) Population genetic structure of black-tailed prairie dogs, a highly interactive species, in fragmented urban habitat. J Mammal 91:326–335
Magle SB, Hunt VM, Vernon M, Crooks KR (2012) Urban wildlife research: past, present, and future. Biol Conserv 155:23–32
McClenaghan LR, Truesdale HD (2002) Genetic structure of endangered Stephens’ kangaroo rat populations in southern California. Southwest Nat 47:539–549
Meshriy MG, Randall JA, Parra L (2011) Kinship associations of a solitary rodent, Dipodomys ingens, at fluctuating population densities. Anim Behav 82:643–650
Munshi-South J, Kharchenko K (2010) Rapid, pervasive genetic differentiation of urban white-footed mouse (Peromyscus leucopus) populations in New York City. Mol Ecol 19:4242–4254
Munshi-South J, Zak Y, Pehek E (2013) Conservation genetics of extremely isolated urban populations of the northern dusky salamander (Desmognathus fuscus) in New York City. PeerJ 1:e64
Munshi-South J, Zolnik CP, Harris SE (2016) Population genomics of the anthropocene: urbanization is negatively associated with genome-wide variation in white-footed mouse populations. Evol Appl 9:546–564
NASA Earth Observatory (2016) http://earthobservatory.nasa.gov/Features/MeasuringVegetation/measuring_vegetation_1.php. Accessed 10 Oct 2013
Noël S, Lapointe F-J (2010) Urban conservation genetics: study of a terrestrial salamander in the city. Biol Conserv 143:2823–2831
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org. Accessed Jan 2016
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237
Reed DH, Hobbs GR (2004) The relationship between population size and temporal variability in population size. Anim Conserv 7:1–8
Riley SPD, Pollinger JP, Sauvajot RM, York EC, Bromley C, Fuller TK, Wayne RK (2006) A Southern California freeway is a physical and social barrier to gene flow in carnivores. Mol Ecol 15:1733–1741
Santonastaso TT, Dubach J, Hauver SA, Graser WH III, Gehrt SD (2012) Microsatellite analysis of raccoon (Procyon lotor) population structure across an extensive metropolitan landscape. J Mammal 93:447–455
Segelbacher G, Höglund J, Storch I (2003) From connectivity to isolation: Genetic consequences of population fragmentation in capercaillie across Europe. Mol Ecol 12:1773–1780
Sikes RS, Gannon WL, The Animal Care and Use Committee of the American Socitey of Mammalogists (2011) Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal 92:235–253
Song XP, Sexton JO, Huang C, Channan S, Townshend JR (2016) Characterizing the magnitude, timing and duration of urban growth from time series of Landsat-based estimates of impervious cover. Remote Sens Environ 175:1–13
Sullivan RM, Best TL (1997) Effects of environment on phenotypic variation and sexual dimorphism in Dipodomys simulans (Rodentia: Heteromyidae). J Mammal 78:798–810
Theobald DM (2001) Land-use dynamics beyond the American urban fringe. Geogr Rev 91:544–564
Theobald DM (2005) Landscape patterns of exurban growth in the USA from 1980 to 2020. Ecol Soc 10:1–27
Vega AM, Estrella RR, Blázquez MC (2007) Characterization of nine microsatellite loci on endemic kangaroo rats Dipodomys simulans peninsularis from southern Baja California Peninsula. Mol Ecol Notes 7:127–129
Yuan F, Bauer ME (2007) Comparison of impervious surface area and normalized difference vegetation index as indicators of surface urban heat island effects in Landsat imagery. Remote Sens Environ 106:375–386
Acknowledgements
Research supported by traineeship to GH from NIH RISE 4 and 5 Grant # R25GM061222-16 and a Grant from T&E Inc. Internships for undergraduate students were supported by USDA NRCT at NMSU and Gavilan College STEM. Thanks to C. Smith, E. Hibbard, D. Price, A. Martinez, J. Cervantes, A. Orenday, J. Medina and R. Garcia for assistance in the field, and the City of Las Cruces, Bureau of Land Management—Las Cruces District and New Mexico State University for allowing this study to occur on their respective properties. Thanks to R. Berg for assistance in screening microsatellites. We thank two anonymous reviewers for comments that improved the manuscript. This material is based upon work while KEM was serving at the National Science Foundation. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hurtado, G., Mabry, K.E. Genetic structure of an abundant small mammal is influenced by low intensity urbanization. Conserv Genet 20, 705–715 (2019). https://doi.org/10.1007/s10592-019-01163-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01163-7