Abstract
Phosphoenolpyruvate carboxylase (PEPC) is a key enzyme in the C4 photosynthetic pathway for the initial fixation of atmospheric CO2 into the four-carbon (C4) acid, oxaloacetate. Here, we report that a vector carrying the intact maize C4 pepc gene was used to transform C3 monocot japonica rice cultivar Tainung 67 (TNG67) which is an important cultivar in Taiwan, and C3 dicot Cleome spinosa which is closely related to the C4 dicot Cleome gynandra, via an Agrobacterium-mediated method. In total, 29 transgenic rice and 4 transgenic C. spinosa plants were obtained. Genomic PCR and southern blot analyses revealed that the maize pepc and selective antibiotic resistant genes were present in almost all randomly-selected transgenic plants but not in wild-type plants. Reverse transcription-PCR showed that maize pepc mRNA was detected in transgenic plants. Western blot analysis confirmed the presence of 110 kDa maize PEPC protein in transgenic plants but not in wild-type plants. Furthermore, higher PEPC enzyme activities were detected in transgenic rice plants than in transgenic C. spinosa plants. Taken together, we clearly demonstrated monocot maize C4 pepc gene is functional in both C3 monocot rice and C3 dicot C. spinosa plants. Two T5 homozygous rice lines, each harboring a single insertion of maize pepc gene, showed 7- to 9.4-fold and 40–54% PEPC enzyme activity as compared to the untransformed rice and maize, respectively. These stable transgenic rice lines will be valuable material for studying the effect of maize PEPC on rice photosynthesis under different conditions.
Similar content being viewed by others
Introduction
Most important crops including rice, wheat, soybean and potato are C3 plants that assimilate atmospheric CO2 directly through the C3 photosynthetic pathway, in which a C3 acid phosphoglycerate is formed via ribulose 1,5-bisphosphate carboxylase (Rubisco) in mesophyll cells. In contrast, a few other important crops such as maize, sugarcane and sorghum use the C4 photosynthetic pathway, in which atmospheric CO2 is first converted to HCO3 − by carbonic anhydrase (CA) and then fixed into the four-carbon (C4) acid, oxaloacetate (OAA), by phosphoenolpyruvate carboxylase (PEPC) in mesophyll cells. OAA is converted to malate or aspartate, and then transported into the neighboring bundle sheath cells where the conventional C3 cycle is used (Edwards 1999; Kajala et al. 2011; Williams et al. 2012). In general, C4 plants are considered more efficient in fixing carbon dioxide with high water and nitrogen use efficiencies, leading to rapid growth rates, higher dry matter production and larger economic yields than C3 plants.
PEPC is a key enzyme in the C4 photosynthetic pathway, catalyzing the initial fixation of atmospheric CO2; therefore, the expression of C4-type pepc gene in C3 rice plants is thought to be a primary event in the establishment of C4 photosynthetic activity in rice to improve the photosynthetic efficiency. Previously, the transformation vector PEPCgenome/pBIH2 carrying the XbaI fragment (8.8 kb) of maize pepc gene (including all exons and introns, promoter and terminator sequences) and two selective antibiotic resistant genes neomycin phosphotransferase II (nptII) and hygromycin phosphotransferase II (hptII) was used to transform two japonica rice cultivars (Kitaake and Nipponbare) via an Agrobacterium-mediated method. Most of the transgenic rice plants generated showed high-level expression of the maize pepc gene and exhibited reduced sensitivity of photosynthesis to O2 inhibition (Ku et al. 1999). Later, the same construct was used to transform indica rice IR68899B using a bombardment method, and the transformants showed enhancement in photosynthesis rate under high temperature (Bandyopadhyay et al. 2007). Now, it is generally agreed that overexpression of a single maize pepc gene encoding PEPC protein or even quadruple transformants overexpressing all four key enzymes in the C4 photosynthetic pathway [i.e., PEPC, pyruvate, orthophosphate dikinase (PPDK), NADP-malate dehydrogenase (NADP-MDH), NADP-malic enzyme (NADP-ME)] into rice plants do not show dramatic improvements in photosynthesis under normal conditions (Taniguchi et al. 2008; Kajala et al. 2011; Miyao et al. 2011). However, a recent study with transgenic wheat plants that overexpressed maize ppdk cDNA, maize pepc cDNA and both showed enhanced maximum daily photosynthetic rates of 4.5, 13.3 and 26.4%, respectively, and enhanced daily accumulated photosynthesis of 6.9, 13.9 and 21.3%, respectively, as compared with the wild-type plants (Zhang et al. 2014). Also, the Fv/Fm of these transgenic wheat lines decreased less than that of the control under high temperature and high light conditions, suggesting that the transgenic wheat plants transport more absorbed light energy to photochemical reactions. The enzyme activities of PPDK and PEPC in these wheat lines were only about twofold and fourfold, respectively, higher than those of the control plants. These expression levels are much lower than those achieved in the transgenic rice plants (Ku et al. 1999; Taniguchi et al. 2008). Whether the differences in expression levels of these maize genes in the C3 rice and wheat affect their photosynthetic performance and other physiological traits under different conditions remain to be investigated.
In this study, an Agrobacterium-mediated method established in our laboratory was adopted to transform the PEPCgenome/pBIH2 vector into the japonica rice cultivar Tainung 67 (TNG67), an important rice cultivar in Taiwan, and a C3 dicot plant Cleome spinosa (Tsai et al. 2012). The genus Cleome is closely related to Arabidopsis; but more interestingly, contains C3 and C4 species. For example, C. spinosa was identified as having C3-type photosynthesis, whereas C. gynandra was identified as having C4-type photosynthesis (Voznesenskaya et al. 2007). In addition, C. spinosa, also known as “spider flower”, is widely used as floricultural plant. Thus, it is a valuable system to choose the C3 plant C. spinosa, which is close to the C4 plant C. gynandra. The genus Cleome was proposed as a model for understanding the differences between C3 and C4 photosynthesis over 10 years ago (Brown et al. 2005), and has been used by a number of groups since then. High levels of maize PEPC protein and enzyme activity were detected in the transformants. Obtaining stable transgenic rice plants with varying expression levels will be useful for further studies, such as analysis of photosynthetic performance under stress conditions.
Materials and methods
Plant material and culture conditions
Seeds of rice (Oryza sativa L. ssp. japonica cultivar TNG67) were used in this study. Dehulled seeds were sterilized by sequential treatment with 70% ethanol for 0.5 min and 1% sodium hypochoride for 10 min, then washed thoroughly with sterile water, and inoculated on NB medium (Sallaud et al. 2003) in the dark for 3 weeks in a 25 °C growth chamber. Calli (embryogenic units) were excised and transferred onto fresh NB medium in a dark growth chamber (25 °C) for 2 weeks before transformation.
Seeds of C. spinosa cultivar Queen were purchased from Know-You Seed Company, Taiwan, and were grown in a greenhouse until maturity. Freshly collected seeds from white flowering plants were sterilized, and germinated on germination medium as previously described (Tsai et al. 2012). In vitro cotyledons were used in transformation experiments.
Agrobacterium-mediated transformation
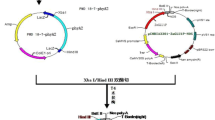
The transformation vector PEPCgenome/pBIH2 (Fig. 1), which contains the 8.8 kb genomic DNA fragment of maize phosphoenolpyruvate carboxylase (PEPC) including its promoter and terminator sequences, and nptII (for kanamycin resistance) and hptII (for hygromycin resistance) two selection marker genes, was transformed into the Agrobacterium tumefaciens strain EHA105 by electroporation (Wang et al. 2015). A single colony from a YEP plate (1% peptone; 1% yeast extract; 0.5% NaCl; 1.5% Bacto-agar) was picked up and inoculated overnight in 5 ml YEP liquid medium supplemented with 50 µg ml−1 kanamycin at 28 °C with shaking at 200 rpm. Bacterial culture (50 µl) was transferred into 50 ml YEP/kanamycin liquid medium supplemented with 100 µM acetosyringone, further incubated overnight at 28 °C with shaking, and the OD600 of Agrobacterium was adjusted to a value of 1.0 by using CCL medium (Sallaud et al. 2003).
Schematic diagram of the T-DNA region of the transformation vector PEPCgenome/pBIH2. Intact maize pepc gene (8.8 kb) including promoter region (from −1212), 10 exons and 9 introns, and terminator region (about 2.5 kb) was inserted between two selective antibiotic resistant genes neomycin phosphotransferase II (NPTII) and hygromycin phosphotransferase II (HPTII). The location of Intron 4 is indicated, which was used for examination of intron-splicing in RT-PCR analysis and Southern blot probe preparation
Detail protocols for rice transformation were as previously described (Sallaud et al. 2003) with modifications. In brief, 50–100 embryogenic calli were co-incubated with 25 ml Agrobacterium for 15 min. The calli were blotted dry on sterilized filter paper, and then 10 calli were transferred on a plate containing co-culture medium (CCS) for 3 days at 25 °C in the dark. After washing, 5–7 calli were transferred onto a plate containing R2S medium (supplemented with 50 mg l−1 hygromycin for selection and 400 mg l−1 cefotaxime to eliminate Agrobacterium) for 14 days in the dark. The brownish calli were transferred onto NBS medium (supplemented with hygromycin and cefotaxime) for 7 days, those that developed brownish globular structures from the surface of callus were picked up by forceps, and inoculated on the same NBS medium for 2 weeks. The round shaped and yellowish calli were transferred into PRAG pre-regeneration medium containing 50 mg l−1 hygromycin and 400 mg l−1 cefotaxime, and incubated at 25 °C growth chamber in the dark for 7 days. The creamy-white calli with a smooth and dry appearance were transferred onto RN regeneration medium containing cefotaxime and hygromycin, incubated at 25 °C in a growth chamber in the dark for 2 days, and then shifted to a 16-h light/8-h dark photoperiod incubator (25 °C) for 3 weeks. Shoots from resistant calli were excised and transferred onto P medium (without antibiotics) for root development in a 16-h light/8-h dark photoperiod incubator (25 °C) for 3 weeks. Plantlets were transferred to soil pots and grown in a greenhouse.
Protocols and media for C. spinosa regarding co-culture, Agrobacterium infection, kanamycin selection and plant regeneration were as previously described (Tsai et al. 2012).
Transgenic plant verification by genomic PCR analysis
Transgenic plant verification was performed with putative transformants and wild-type plants by PCR analysis. Total genomic DNA was extracted from green leaves using the CTAB method (Wilkie 1997). PCR was carried out with the following primer sets: F-1201 (5′-TCT AGA GAT GTA ATG GTG TTA GGA CAC-3′) and R153 (5′-TCG GAG ACC TTG CCT GGG AC-3′) for amplification of 1385-bp-long DNA fragment corresponding to the promoter region and partial coding sequences of maize pepc in the transforming vector PEPCgenome/pBIH2; HPTII_F (5′-ATG AAA AAG CCT GAA CTC ACC-3′) and HPTII_R (5′-CCT TTG CCC TCG GAC GAG TG-3′) for amplification of the 1021-bp-long DNA fragment corresponding to the selection marker hygromycin resistant gene (hptII); and Kan-F (5′-ATG ATT GAA CAA GAT GGA-3′) and Kan-R (5′-TCA GAA GAA CTC GTC AAG-3′) for amplification of the 795-bp-long DNA fragment corresponding to the full-length kanamycin resistant gene (nptII). PCR reactions were performed in a volume of 20 µl under the following conditions: (a) denaturation for 5 min at 94 °C; (b) 30 cycles of 0.5 min at 94 °C, 0.5 min at 55 °C, and 1 min at 72 °C; and (c) extension for 15 min at 72 °C. Following amplification, 10 µl PCR products were analyzed on a 1% agarose gel.
Transgene expression analysis by reverse transcription-PCR (RT-PCR)
Total RNA from green leaves of wild-type and transgenic plants was isolated by TRIzol reagent as previously described (Wang et al. 2009), treated with DNase I and 400 ng of total RNA was used to conduct RT-PCR analysis by using one-step RT-PCR kit (Qiagen). Primers ZmPEPC-f2 (5′-CAG AAC CGA TGA AAT CAG G-3′) and ZmPEPC-r2 (5′-CAG CTC TTC AAT CTG ATC G-3′) were located in the flanking regions of intron 4 (449 bp) in maize PEPC genomic DNA (Fig. 1). The size of the unspliced product is 753 bp, while the size of the spliced product is 304 bp. For internal control in rice, primers OsActin1-F (5′-TCC ATC TTG GCA TCT CTC AG-3′) and OsActin1-R (5′-TAC CCT CAT CAG GCA TCT G-3′) were employed to amplify the actin fragment of 337 bp; and for internal control in C. spinosa, primers CsEF1-f1 (5′-ATC AAC ATT GTG GTC ATT GG-3′) and CsEF1-r1 (5′-TTG ATC TGG TCA AGA GCC-3′) were employed to amplify the elongation factor 1 fragment of 650 bp. Reverse transcription was carried out at 50 °C for 30 min. Initial PCR activation was carried out at 94 °C for 15 min. DNA amplification was carried out for 35 cycles (denaturation at 94 °C for 0.5 min, annealing at 55 °C for 0.5 min, extension at 72 °C for 1 min). Final extension was performed at 72 °C for 10 min, and the PCR product was stored overnight at 15 °C in a PCR machine (PTC-200, Peltier Thermal Cycler). Following amplification, PCR products were analyzed on a 1% agarose gel.
Southern blot analysis
For wild-type and transgenic rice plants, 15 µg of plant DNA was digested overnight at 37 °C with restriction enzyme EcoRI. After electrophoresis on 0.8% agarose gel in TAE buffer, the gel was washed, denatured, and transferred onto membrane as previously described (Tsai et al. 2012). To prepare the non-radioactive PCR DIG probe (Roche Applied Science, Germany), equal amounts (20 pmol) of primers ZmPEPC-f2 and ZmPEPC-r2 were used to amplify the foreign gene maize pepc DNA fragment (753 bp) from the expression vector PEPCgenome/pBIH2 (1 µg) and the reaction mixture as previously described (Tsai et al. 2012). For PCR amplification of the probe, hybridization, and detection were as previously described (Tsai et al. 2012).
For wild-type and transgenic C. spinosa plants, 15 µg of plant DNA was used for overnight digestion at 37 °C with restriction enzyme HindIII. After electrophoresis and transfer onto membrane, DIG-labelled PCR products from two gene-specific primer sets, Kan-F and Kan-R for amplification of the 795-bp-long DNA fragment corresponding to the full-length selection marker nptII gene, and ZmPEPC-f2 and ZmPEPC-r2 for the 753-bp-long DNA fragment corresponding to the maize pepc gene were used as probes in hybridization experiments. Detailed protocols were described previously (Tsai et al. 2012).
Seedling assay for antibiotic resistance
For rice plants, dehulled seeds were sterilized by sequential treatment with 70% ethanol and 1% sodium hypochoride for 10 min, washed thoroughly with sterile water, and then inoculated on germination medium containing MS salts (Murashige and Skoog 1962), 3% sucrose, 0.8% Bacto-agar, 50 mg l−1 hygromycin. The cultures were incubated at 25 °C under 16-h illumination for 2 weeks. Seedlings that displayed restricted growth, had pale green or white aerial parts, and showed inhibition of main root extension were considered to be hygromycin sensitive, whereas seedlings with healthy development of leaves and roots were considered to be hygromycin resistant.
Seedling assay for transgenic C. spinosa plants resistant to 100 mg l−1 kanamycin was as previously described (Tsai et al. 2012).
Western blot analysis and assay of PEPC activity
Protein extraction, gel electrophoresis, western blot analysis, and assay of PEPC activity were as previously described (Ku et al. 1999). Briefly, 0.1 g leaf tissue from wild-type and transgenic plants was harvested and quickly ground in 1.5 ml extraction buffer (50 mM Tris–HCl, pH 7.0; 10 mM MgCl2; 1 mM EDTA; 5 mM dithiothreitol; 5% polyvinylpolypyrrolidone; 10% glycerol) using a cold pestle and mortar. Leaf extract was centrifuged at 15,000g for 10 min, and the supernatant was used immediately for assay of PEPC. PEPC activity was assayed spectrophotometrically at room temperature in a mixture [50 mM Tricine–KOH, pH 8.0; 0.1 mM EDTA; 1 mM dithiothreitol; 10 mM MgCl2; 10 mM NaHCO3; 3 U NAD-malate dehydrogenase; 0.2 mM NADH; 0.01 ml protein extract (0.03 mg protein)]. The reaction was initiated by adding phosphoenolpyruvate to a final concentration of 2 mM. For western blot analysis, 15 µg of total soluble protein from leaf extract was electrophoretically separated on 10% SDS–polyacrylamide gel, and transferred onto a nitrocellulose membrane. The blot was incubated with anti-PEPC antibody, then visualized by incubation with alkaline phosphatase-conjugated goat anti-rabbit IgG followed by color development with BCIP/NBT.
Results
Isolation and confirmation of putative transgenic plants from rice and C. spinosa
As shown in Fig. 1, the transformation vector PEPCgenome/pBIH2 carries both kanamycin and hygromycin selection marker genes. For rice, from a total of 819 seed-derived embryogenic calli, 30 putative transgenic plants were obtained from selection medium containing 50 mg l−1 hygromycin. To avoid redundant transgenic plants, only a single shoot was excised from each callus. Among the 30 putative transgenic plants, one seedling grown in the greenhouse was infected by microorganisms and finally died before further verification. Schematic representation of the typical protocol is shown in Fig. 2.
Agrobacterium-mediated transformation was used to generate transgenic rice plants carrying the C4-specific pepc gene from maize. a Hygromycin resistant calli were proliferated in the selection medium. b Green shoots were initiated from resistant calli. Only one shoot was excised from each callus from further plant regeneration. c The putative transgenic line was grown in test tube. d Putative transgenic plants were transferred into pots and grown in a greenhouse
Using the protocols of plant regeneration and transformation for C. spinosa we previously developed (Tsai et al. 2012), four putative transgenic plants were obtained from a total of 164 cotyledon explants using selection medium containing 50 mg l−1 kanamycin, and then grown in a greenhouse (Fig. 3).
Agrobacterium-mediated transformation was used to generate transgenic Cleome spinosa plants carrying the C4-specific pepc gene from maize. Cotyledon explants were excised from the seedlings grown in vitro, infection with A. tumefaciens strain EHA105 carrying the PEPCgenome/pBIH2 vector, and incubated on selection medium containing 50 mg l−1 kanamycin and 200 mg l−1 timentin. The rooted plantlets were transferred to potting soil. a Transgenic plant at the young stage. b Flowers were observed in a transgenic plant. c Enlarged flower showing plenty of narrow capsules
To verify the rice transformants, we isolated genomic DNA from leaves of plantlets and PCR analysis was carried out using specific primers for pepc (Fig. 4a) or the selection marker hygromycin (hptII) gene (Fig. 4b). No PCR product was detected in wild-type rice (WT). Among the 29 putative transgenic plants, 28 transgenic plants carried both transgenes, as revealed by the presence of two unique PCR bands of 1385 bp (pepc; Fig. 4a) and 1021 bp (hptII; Fig. 4b). One transgenic line (No. 11) is considered to be a false positive line, since the hptII selection marker was detected in its genome and thus was able to grow in 50 mg l−1 hygromycin-containing media R2S and NBS during selection; however, the foreign maize genomic pepc DNA fragment was not detected, probably due to incomplete T-DNA integration in Agrobacterium infection. Loss of one of the two transgenes within the same T-DNA is a frequent event, and has been reported in a range of plants including wheat (Wu et al. 2006), rice (Kim et al. 2003), and our recent studies in Bidens pilosa (Wang et al. 2012) and Cleome spinosa (Tsai et al. 2012). Similarly, to verify transgenic C. spinosa plants, genomic DNA was isolated and PCR analysis was carried out using specific primers for pepc (Fig. 4c), and selection marker gene hptII (Fig. 4d) or kanamycin (nptII; Fig. 4e). No PCR product was detected in the wild-type C. spinosa (WT), while unique PCR bands of 1385 bp (pepc; Fig. 4c), 1021 bp (hptII; Fig. 4d) and 795 bp (nptII; Fig. 4e) were detected in all 4 transgenic lines obtained when specific primers were employed.
Confirmation of transgenes in transgenic rice and Cleome spinosa plants by genomic PCR analysis. a, b Genomic DNA was isolated from wild-type (WT) and 29 putative transgenic rice plants, 200 ng of each DNA sample was used for PCR in the presence of pepc-specific primers F-1201 and R153 (a) or selection marker hptII-specific primers HPT_F and HPT_R (b). The size of PCR product in panel a is 1385 bp, whereas in panel b it is 1021 bp. Plasmid PEPCgenome/pBIH2 was used as positive control (lane P). Molecular sizes are indicated by arrows. c–e Genomic DNA was isolated from wild-type (WT) and four putative transgenic C. spinosa plants, and PCR was carried out using pepc-specific (1383 bp; c), hptII-specific (1021 bp; d) or nptII-specific (795 bp; e) primers
C4 maize pepc gene expression in C3 transgenic plants
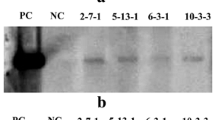
The transformation vector PEPCgenome/pBIH2 carries a maize (a C4 plant) pepc genomic fragment (8.8 kb) containing promoter, all introns and exons, and terminator sequences (Fig. 1). To investigate whether the C4 maize pepc gene can be expressed and is functionally active in the C3 monocot rice and the C3 dicot C. spinosa plants, the first step was to confirm whether the C4 maize pepc gene can be transcribed into pre-mRNA and then pre-mRNA can be spliced into mature mRNA. Nucleotide sequence analysis revealed that the length of the maize pepc gene including 10 exons and 9 introns is 5022 bp (from the first nucleotide of start codon ATG to the last nucleotide of stop codon TAG), and the length of its cDNA is 2913 bp (Matsuoka and Minami 1989). At the very beginning, we did not obtain any PCR product for full-length cDNA when forward and reverse primers for the pepc gene were employed, probably due to its large size for amplification. Figure 5 shows the splicing of Intron 4 (449 bp in length), which is the second largest intron in the pepc genomic sequence. The forward primer ZmPEPC-f2 is located at Exon 3, whereas the reverse primer ZmPEPC-r2 is located at Exon 5. If the transformed maize C4 pepc gene is also functionally transcribed and spliced into mRNA in C3 transgenic plants, the expected PCR product should be 304 bp after splicing of Intron 4, whereas the unspliced PCR product should be 753 bp. Results from RT-PCR analysis revealed that no PCR product was detected in RNA samples isolated from wild-type rice (Fig. 5a) or wild-type C. spinosa (Fig. 5c), indicating the specificity and reliability of the assay. In contrast, a unique band of 0.3 kb was detected in all 14 random-selected transgenic rice plants examined (Fig. 5a), indicating that Intron 4 (449 bp) is indeed spliced out to form mature mRNA. In other words, maize C4 pepc gene can also be transcribed and spliced into mature mRNA in C3 monocot rice. The transformation vector PEPCgenome/pBIH2 was used as a positive control, a unique PCR band of 753 bp was detected (Fig. 5). This is the expected size if Intron 4 has not been spliced. For transgenic C. spinosa plants, a unique band of 0.3 kb was detected in 3 (i.e., #2, #6, #14) out of 4 transgenic plants (Fig. 5c), indicating that maize C4 pepc gene can also be transcribed and spliced correctly into mature mRNA in C3 dicot C. spinosa plants. No PCR product was detected in transgenic plant #8 (Fig. 5c). The reason is not clear yet.
Examination of accuracy of intron-splicing in transgenic rice (a, b) and Cleome spinosa (c, d) plants by reverse transcription-PCR (RT-PCR) analysis. Total RNA was isolated from leaf of wild-type (WT) and random-selected 14 transgenic rice plants (a), or from wild-type (WT) and four transgenic C. spinosa plants (c), 400 ng RNA from each sample was used for RT-PCR in the presence of primers ZmPEPC-f2 and ZmPEPC-r2, which are the flanking sequences of the Intron 4 in the maize pepc gene. The expected size of the PCR product from genomic DNA is 753 bp, whereas the expected size of PCR product from the mRNA is 304 bp (a, c). As an internal control in rice, OsActin1 was amplified, yielding a product of 337 bp (b). For internal control in C. spinosa, CsEF1 was amplified, yielding a product of 650 bp (d). Plasmid PEPCgenome/pBIH2 carrying maize pepc genomic DNA was used as a positive control (lanes P). RT-PCR without template DNA was used as a negative control (lanes N)
Presence of single to multiple copies of maize pepc gene of transgenic plants
To estimate the copy number in each transgenic rice plant, genomic DNA from wild-type and 8 random-selected transgenic plants was digested with EcoRI and the DNA fragments were resolved electrophoretically. To prepare the PCR probe, the same primers, ZmPEPC-f2 and ZmPEPC-r2, were used to amplify the 753 bp DNA fragment in the maize pepc genomic sequence from the transformation vector PEPCgenome/pBIH2. No internal EcoRI restriction site was found in the probe. No hybridization band was detected in the wild-type DNA sample (Fig. 6). The hybridization bands in all samples were different, indicating that all of the 8 transgenic plants were independent integration events. A single copy of the maize pepc gene was integrated into the genome of transgenic plants #3, #17 and #28, two copies of the maize gene were detected in transgenic plants #1, #2 and #27, and 4 to 5 copies of the maize gene were detected in transgenic plants #4 and #26 (Fig. 6).
For transgenic C. spinosa plants, genomic DNA from wild-type and 4 transgenic plants was digested with HindIII, and probed with the selection marker gene nptII (795 bp) or the same maize pepc fragment (753 bp). No internal HindIII restriction site was found in the selection marker nptII gene. No hybridization band was detected in the wild-type sample in either Southern blot (Fig. 7). Again, the hybridization bands in all samples were different, indicating that all of the 4 transgenic plants were independent integration events. For the Southern blot probed with the selection marker nptII, multiple copies were detected in transgenic plant #2, and 3 copies were detected in transgenic plants #6 and #8 (Fig. 7a). This is consistent with the results obtained from the Southern blot probed with the maize pepc fragment (753 bp), except that only one hybridization band was detected in transgenic plant #8 (Fig. 7b). For transgenic plant #14, although the hybridization signal was detected by both probes, the size of the hybridization band was quite large and the signal was quite thick. We, therefore, believe that multiple copies of the transgene were inserted into the host chromosomal genome in this transgenic plant.
Transgenes are stably transmitted into their progenies
To demonstrate the stability and inheritance of the transgenes, three transgenic rice plants (#3, #17, #28), each harboring a single copy of the transgene as revealed by Southern blot (Fig. 6), were transferred to a greenhouse until maturation. One of transgenic plants (#28) was infected by microorganisms. Seeds collected from selfed T0 plants (#3, #17) were sterilized and germinated on selection medium containing 50 mg l−1 hygromycin. The number of resistant seedlings (58) to sensitive seedlings (21) in transgenic plant #3 and resistant seedlings (61) to sensitive seedlings (19) in transgenic plant #17 showed a Mendelian segregation ratio of R:S = 3:1. Then, nine and seven T1 resistant seedlings from transgenic plants #3 and #17, respectively, were transferred into the greenhouse. Seedling assay was carried out using approximately 60 T2 seeds. From total five samples (three from T2 transgenic #3 seeds and two from T2 transgenic #17 seeds) we examined, two homozygous T2 seeds (i.e., #3-7 and #17-3) were identified, as all the 60 T2 seedlings from T1 transgenic #3-7 and #17-3 plants showed a resistant phenotype in hygromycin-containing medium. Since homozygous lines #3-7 and #17-3 representing transgenic plants #3 and #17, respectively, were obtained; therefore, we stopped screening the rest (11) of T2 seeds. Homozygous plants were transferred into pots and grown up to the T5 generation. As an example, all T4 seedlings from transgenic lines (#3 and #17) showed a resistant phenotype in hygromycin-containing medium (Fig. 8). Thus, we concluded that a single copy of the foreign DNA cassette was inserted into the nuclear chromosome in transgenic plants #3 and #17, and the transgenes were transmitted in a Mendelian manner and stably inherited from their transgenic parents into their progenies.
For transgenic C. spinosa plants, multiple copies of the maize PEPC gene were detected as revealed by Southern blot analysis (Fig. 7), all 4 transgenic plants we obtained were transferred into pots and grown in a greenhouse until maturation. Seeds collected from selfed T0 plants were sterilized and then germinated on kanamycin-containing medium. As expected, all T1 seedlings from all 4 transgenic plants showed resistant phenotype (data not shown), since T1 sensitive seedlings are relatively rare from a limited number of seeds under assay. No effort was made to screen for homozygous lines from the plants, as it may take quite a long time to breed a homozygous line carrying multiple copies of transgenes.
Detection of maize PEPC protein and PEPC enzyme activity in transgenic plants
To evaluate the presence and function of maize PEPC protein in the transgenic plants, total protein was extracted from leaf tissue and subjected to western blot analysis. Two individual T5 resistant plants from each of the single homozygous lines of transgenic rice (i.e., #3-1, #3-2, #17-1, #17-2) were selected from hygromycin-containing medium and grown in a greenhouse, and the expression of the maize pepc gene in transgenic rice plants was confirmed by the presence of the 110-kDa protein specific for the maize enzyme (Fig. 9a). For the transgenic C. spinosa plants, 2 individual T1 resistant plants from 3 transgenic lines (i.e., #2-1, #2-2, #6-1, #6-2, #14-1, #14-2) were selected from kanamycin-containing medium and grown in a greenhouse, and expression of the pepc gene in transgenic C. spinosa plants was also confirmed by the presence of the 110-kDa protein specific for the maize enzyme (Fig. 9b). It is well documented that the maize PEPC enzyme is composed of 970 amino acid residues with a molecular mass of 109,408 Da (Matsuoka and Minami 1989).
Next, PEPC enzyme activity was measured to examine the function of the maize pepc gene in transgenic plants. As expected, a minimal or background level of PEPC activity was detected in the leaves of wild-type rice (average = 56 µmol mg chl−1 h−1; Fig. 10a) or C. spinosa (7 µmol mg chl−1 h−1; Fig. 10b). Average PEPC activity in the leaves of 2 T5 plants from transgenic rice line 3 (namely, #3-1 and #3-2) was 529 µmol mg chl−1 h−1, which is 9.4-fold higher than that of wild-type rice and is close to 54% of the maize activity (Fig. 10a). For another transgenic line, #17, average PEPC activity in the leaves of two T5 plants (namely, #17-1 and #17-2) was 397 µmol mg chl−1 h−1, which is sevenfold higher than that of wild-type rice and is close to 40% of the maize activity (Fig. 10a). For transgenic C. spinosa plants (Fig. 10b), average PEPC activity in the leaves of two individual T1 plants from transgenic line #2 (namely, #2-1 and #2-2) was 152 µmol mg chl−1 h−1, which is much lower than those of transgenic rice plants obtained. Average PEPC activity in transgenic lines #14-1 and #14-2 was 100 µmol mg chl−1 h−1. Similarly, a low PEPC activity was detected in the leaves of transgenic line #6, which is consistent with the very faint band observed in western blot analysis (Fig. 9b).
Discussion
In this study, the vector PEPCgenome/pBIH2 carrying intact maize pepc genomic DNA fragment and two selective antibiotic resistant genes (nptII and hptII) was used to transform the monocot rice and dicot C. spinosa plants by Agrobacterium-mediated method. For rice, both pepc-specific and hptII-specific fragments were detected in 28 transgenic plants regenerated from a total of 819 seed-derived embryogenic calli (Fig. 4a, b). Thus, the transformation efficiency for rice was 3.4%. For C. spinosa, 4 transgenic plants were obtained from a total of 164 cotyledon explants, and pepc-specific, hptII-specific and nptII-specific fragments were detected in all these plants (Fig. 4b, c, e). Thus, a slightly lower transformation efficiency of 2.4% was achieved for C. spinosa. Although the transformation efficiency in rice is much lower than reported previously (75–95%; Sallaud et al. 2003), the transformation efficiency in C. spinosa is similar to our previous report (3.5%; Tsai et al. 2012). The large variation in rice transformation efficiency could be attributed to the use of different cultivars. Relative to recalcitrant indica rice cultivars, japonica rice cultivars exhibit much higher transformation efficiency. Though TNG67 is classified as a japonica rice it has some indica rice background (Dalrymple 1986) and this may account for its lower efficiency. More importantly, using the rigorous criteria during selection, all transformants we examined are independent lines as revealed by Southern blot analysis (Figs. 6, 7). Redundant transformants should be eliminated before analyzing any results for the transgene. No apparent differences in phenotype were observed in any of the transgenic plants as compared to their wild-type parents. This result is similar to other reports that over 90% transgenic rice plants carrying the same construct in different cultivars exhibited normal phenotype as the wild-type origin (Ku et al. 1999; Bandyopadhyay et al. 2007).
Two T5 homozygous lines (i.e., #3 and #17), each harboring a single insertion of maize pepc gene, have been identified from transgenic japonica rice cultivar TNG67. About 7- to 9.4-fold or 40–54% of PEPC enzyme activities were measured as compared to untransformed wild-type rice or wild-type maize, respectively (Fig. 10). Previously, PEPC enzyme activities onefold to threefold higher than wild-type maize were detected in two transgenic japonica rice cultivars Kitaake and Nipponbare using the same construct (Ku et al. 1999), and progenies from several generations of these transformants were used studying physiological and metabolic enzyme activities during the flowering stage (Xia and Cao 2013; Chen et al. 2014; Qian et al. 2015). These studies revealed an enhanced drought tolerance of transgenic rice plants expressing high levels of maize PEPC by more efficiently scavenging reactive oxyradicals. Moreover, the same construct has been used to transform the indica rice line IR68899B and transformants showed enhanced photosynthesis rate under high temperature conditions (Bandyopadhyay et al. 2007). Most recently, transgenic wheat plants were transformed with pepc cDNA and showed about fourfold higher PEPC activity, relative to the wild-type wheat (Zhang et al. 2014). The transgenic wheat plants exhibited enhanced photosynthetic performance and stress tolerance. No evaluation has been made in these studies whether the differences in expression level affect the photosynthetic performance and stress tolerance. Thus, we believe that our transgenic rice lines in the japonica cultivar TNG67, a popular commercial cultivar, with varying expression levels are valuable material for studying physiological responses during the vegetative and reproductive stages or investigating the photosynthetic performance under various stress conditions, since dramatic differences may occur between different cultivars under the same conditions. Coexpression of one or more key genes from the C4 photosynthetic pathway into our transgenic lines carrying maize pepc gene is also an interesting topic to be explored.
References
Bandyopadhyay A, Datta K, Zhang J, Yang W, Raychaudhuri S, Miyao M, Datta SK (2007) Enhanced photosynthesis rate in genetically indica rice expressing pepc gene cloned from maize. Plant Sci 172:1204–1209
Brown NJ, Parsley K, Hibberd JM (2005) The future of C4 research—maize, Flaveria or Cleome? Trends Plant Sci 10:215–221
Chen B, Li X, Huo K, Wei X, Dai C, Lv C (2014) Promotion of photosynthesis in transgenic rice over-expressing of maize C4 phosphoenolpyruvate carboxylase gene by nitric oxide donors. J Plant Physiol 17:458–466
Dalrymple DG (1986) Development and spread of high-yielding rice varieties in developing countries. Bureau for Science and Technology, Agency for International Development. Metrotec, Inc, Washington, D.C., p 124
Edwards G (1999) Tuning up crop photosynthesis. Nat Biotechnol 17:22–23
Kajala K, Covshoff S, Karki S, Woodfield H, Tolley BJ, Dionora MJA, Mogul RT, Mabilangan AE, Danila FR, Hibberd JM, Quick WP (2011) Strategies for engineering a two-celled C4 photosynthetic pathway into rice. J Exp Bot 62:3001–3010
Kim SR, Lee J, Jun SH, Park S, Kang HG, Kwon S, An G (2003) Transgene structures in T-DNA-inserted rice plants. Plant Mol Biol 52:761–773
Ku MSB, Agarie S, Nomura M, Fukayama H, Tsuchida H, Ono K, Hirose S, Toki S, Miyao M, Matsuoka M (1999) High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nat Biotechnol 17:76–80
Matsuoka M, Minami E-i (1989) Complete structure of the gene for phosphoenolpyruvate carboxylase from maize. Eur J Biochem 181:593–598
Miyao M, Masumoto C, Miyazawa S-I, Fukayama H (2011) Lessons from engineering a single-cell C4 photosynthetic pathway into rice. J Exp Bot 62:3021–3029
Murashige T, Skoog F (1962) A revised medium for the rapid growth and bioassays with tobacco cultures. Physiol Plant 15:473–497
Qian B, Li X, Liu X, Chen P, Ren C, Dai C (2015) Enhanced drought tolerance in transgenic rice over-expressing of maize C4 phosphoenolpyruvate carboxylase gene via NO and Ca2+. J Plant Physiol 175:9–20
Sallaud C, Meynard D, van Boxtel J, Gay C, Bès M, Brizard JP, Larmande P, Ortega D, Raynal M, Portefaix M, Ouwerkerk PBF, Rueb S, Delseny M, Guiderdoni E (2003) High efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor Appl Genet 106:1396–1408
Taniguchi Y, Okawa H, Masumoto C, Fukuda T, Tamai T, Lee K, Sudoh S, Tsuchida H, Sasaki H, Fukuyama H, Miyao M (2008) Overproduction of C4 photosythetic enzymes in transgenic rice plants: an approach to introduce the C4-like photosynthetic pathway into rice. J Exp Bot 59:1799–1809
Tsai YT, Chen PY, To KY (2012) Plant regeneration and stable transformation in the floricultural plant Cleome spinosa, a C3 plant closely related to the C4 plant C. gynandra. Plant Cell Rep 31:1189–1198
Voznesenskaya EV, Koteyeva NK, Chuong SDX, Ivanova AN, Barroca J, Craven LA, Edwards GE (2007) Physiology, anatomical and biochemical characterization of photosynthetic types in genus Cleome (Cleomaceae). Funct Plant Biol 34:247–267
Wang HM, Yin WC, Wang CK, To KY (2009) Isolation of functional RNA from different tissues of tomato suitable for developmental profiling by microarray analysis. Bot Stud 50:115–125
Wang CK, Hsu SY, Chen PY, To KY (2012) Transformation and characterization of transgenic Bidens pilosa L. Plant Cell Tissue Organ Cult 109:457–464
Wang CK, Chin YC, Lin CY, Chen PY, To KY (2015) Transforming the snapdragon aurone biosynthetic genes into petunia alters coloration patterns in transgenic flowers. Adv Biosci Biotechnol 6:702–722
Wilkie S (1997) Isolation of total genomic DNA. In: Clark MS (ed) Plant molecular biology—a laboratory manual. Springer, Berlin, pp 3–15
Williams BP, Aubry S, Hibberd JM (2012) Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci 17:213–220
Wu H, Sparks CA, Jones HD (2006) Characterization of T-DNA loci and vector backbone sequences in transgenic wheat produced by Agrobacterium-mediated transformation. Mol Breed 18:195–208
Xia L, Cao W (2013) Physiological and metabolic enzymes activity changes in transgenic rice plants with increased phoshoenolpyruvate carboxylase activity during the flowering stage. Acta Physiol Plant 35:1503–1512
Zhang HF, Xu WG, Wang HW, Hu L, Li Y, Qi XL, Zhang L, Li CX, Hua X (2014) Pyramiding expression of maize genes encoding phosphoenolpyruvate carboxylase (PEPC) and pyruvate orthophosphate dikinase (PPDK) synergistically improve the photosynthetic characteristics of transgenic wheat. Protoplasma 251:1163–1173
Acknowledgements
The authors thank Miranda Loney for editing of the manuscript and Director Wen-Hsiung Li (Biodiversity Research Center, Academia Sinica) for initiating and supporting the research project. We also thank Ku-Feng Lin, Manager of Greenhouse Core Facilities of Academia Sinica, for providing and maintaining growth chambers. This study was financially supported by Academia Sinica (AS-102-SS-A13) and the Innovative Translational Agricultural Research Program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, PY., Tsai, YT., Ng, CY. et al. Transformation and characterization of transgenic rice and Cleome spinosa plants carrying the maize phosphoenolpyruvate carboxylase genomic DNA. Plant Cell Tiss Organ Cult 128, 509–519 (2017). https://doi.org/10.1007/s11240-016-1128-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-016-1128-9