Abstract
This paper presents the first geographical record of the needle nematode Longidorus artemisiae Rubtsova, Chizhov & Subbotin, 1999 outside Russia. This species was found in Poland near the city of Skierniewice in association with nettle (Urtica dioica L.). Morphometric and morphological data are provided, including the first description of juveniles of this species. Nematodes of the Polish population differ from the type-population in Russia in possessing a thicker body (lower ‘a’ index) in both sexes; males having a longer body and longer spicules; different sex ratio (1:2 in Polish population vs 1:1 in the type-population) and somewhat less expanded lips. Molecularly, the Polish population was characterised by sequencing D2-D3 28S rDNA and ITS1 markers. Additionally, new data on these two markers are provided for another species, Longidorus juglandicola Lišková, Robbins & Brown, 1997, obtained from topotype specimens from Slovakia. Surprisingly, despite the high morphological similarity of these two species, analysis of their phylogenetic position did not show close phylogenetic relation and several other species (less similar in general morphology) appeared more closely related to both L. artemisiae and L. juglandicola.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Longidorus Micoletzky, 1927 consists of obligatory plant ectoparasites, many of which are of economic importance as plant pests. This importance is further augmented by the fact, that eight species of the genus are known as vectors of viruses of the genus Nepovirus (Taylor & Brown, 1997). Additionally, Longidorus is rich in species (158 nominal species according to Peneva et al., 2013), which are discriminated mainly on the basis of morphology and morphometrics. However, this approach is complicated by high levels of intraspecific variability in morphometrics and minor interspecific differences leading to substantial overlap among species and increased potential for misidentification (Gutiérrez-Gutiérrez et al., 2013).
To date, 15 species of the genus Longidorus have previously been reported from Poland (Kornobis & Peneva, 2011; Kornobis 2012). During a survey of the occurrence of the nematodes of the family Longidoridae in Poland, a previously non-recorded species Longidorus artemisiae Rubtsova, Chizhov & Subbotin, 1999 was found. This species was described from Russia on the basis of morphology and morphometrics of adult specimens. Subsequently, Rubtsova et al. (2005) obtained a partial sequence of the D2 domain of 28S rRNA gene from paratype specimens and Subbotin et al. (2014) obtained sequences for D2-D3 domains of 28S rRNA gene and partial sequences of the 18S rRNA gene from other Russian populations and assessed the phylogenetic position of this species. Here we present first record of L. artemisiae in Poland. This population was, however, characterised by large intra-specific differences compared to the population from Russia. This paper provides details on the morphology and morphometrics of the Polish population of L. artemisaie including the description of juveniles and sequences of D2-D3 domain of 28S rRNA gene and ITS1.
Additionally, differences between the type-population from Russia and the population from Poland made the latter somewhat similar to another species, Longidorus juglandicola Lišková, Robbins & Brown, 1997. Longidorus juglandicola was described from Slovakia (Lišková et al., 1997) from the rhizosphere of the walnut (Juglans regia L.) on the basis of morphology and morphometrics of adult and juvenile specimens. Thereafter records of L. juglandicola distribution come only from several localities in Serbia (Barsi & Lamberti, 2002; Krnjaic et al., 2002). As no data on molecular markers of L. juglandicola were available, sequences from specimens of the type-population of L. juglandicola collected at the type-locality Sorozka (Slovakia) in 2015 are presented together with data on phylogenetic relationships of both species.
Materials and methods
A total of 925 soil samples were taken during a survey of the occurrence of longidorid nematodes in Poland. In Slovakia, a soil sample was taken from the type-locality of L. juglandicola (Sorozka, a hill in eastern Slovakia, see Lišková et al., 1997) in May 2015. The soil sample was taken from the rhizosphere of the same walnut tree (Juglans regia L.) in the type-locality. Nematodes were extracted using decant and sieving method (Brown & Boag, 1988), fixation for molecular study in DESS (Yoder et al., 2006); the remaining specimens were heat-killed and fixed in TAF (Courtney et al., 1955). For study of morphology and morphometrics of L. artemisiae, specimens were transferred to glycerol as described by Seinhorst (1959). Microscopic slides were made using paraffin ring method and fiber glass to support the coverslip. Observations, measurements and photographs were made using Leica DM5000 microscope. From DESS fixed specimens temporary mounts were made with specimens of both species and photographs illustrating morphology were made (not presented) to retain possibility of checking the morphology of the specimens if necessary. All measurements are in micrometres and are given as the range followed by the means in parentheses.
Subsequently, temporary mounts were dismantled and genomic DNA was isolated from individual specimens using a QIAamp DNA Micro Kit (Qiagen, Hilden, Germany) according to the manufacturer’s a protocol. DNA concentration was measured using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). Amplification and sequencing of the ITS1 rDNA region was performed using primers rDNA2 (Vrain et al., 1992) and rDNA58S (Cherry et al., 1997); amplification of D2-D3 28S rDNA using primers D2A and D3B (Nunn, 1992). PCR conditions were as follows: 95 °C for 5 min, 35 cycles at 94 °C for 45 s, 59 °C for 1 min, 72 °C for 1 min 30 s and a final extension at 72 °C for 10 min (ITS1) and 94 °C for 4 min, 37 cycles at 94 °C for 30 s, 55 °C for 40 s, 72 °C for 1 min and a final extension at 72 °C for 8 min for 28S rDNA. For D2-D3 28S rDNA PCR amplicons were directly sequenced, for ITS1 PCR amplicons were cloned into pCRTM4-TOPO®vector (TOPO®TA Cloning® Kit for Sequencing, Invitrogen) and used to transform into One Shot® TOP10 competent cells for further sequencing. Sequencing was performed by Genomed (Warsaw, Poland).
The newly-generated sequences were subjected to BLAST search to confirm their nematode origins and subsequently deposited under accession numbers KX137849 and KX137850 for D2-D3 28S rDNA of L. artemisiae and L. juglandicola, respectively. Partial 18S-ITS1-5.8S partial sequences were deposited under numbers KX192397–KX192399, KX192400 and KX192395–KX192396 for L. artemisiae and L. juglandicola, respectively. Sequences were aligned using ClustalW (Thompson et al., 1994) implemented in MEGA6 (Tamura et al., 2013). For phylogenetic analysis, appropriate base substitution model was chosen using jModelTest 2.1.8 (Darriba et al., 2012) under Akaike information criteria. Sequence dataset was analysed with Bayesian inference (BI) in MrBayes 3.2 (Ronquist et al., 2012) under GTR+I+G model (for both markers). Analysis was initiated from random tree and run with four chains for 500,000 and 1,000,000 generations for D2-D3 28S rDNA and ITS1, respectively. Trees were sampled every 100 generations, ‘burn-in’ samples (25%) were discarded and convergence evaluated, and the remaining samples were retained for further analysis. The topologies were used to generate a 50% majority rule consensus tree. Trees were visualised in FigTree (http://tree.bio.ed.ac.uk/software/figtree/), minor graphic changes were made to improve clearness of data presentation.
Family Longidoridae Thorne, 1935
Genus Longidorus Micoletzky, 1922
Longidorus artemisiae Rubtsova, Chizhov & Subbotin, 1999
Host: Urtica dioica L. (Urticaceae).
Locality: Near the city of Skierniewice (51.9662N, 20.2482E), Poland.
Prevalence: Only in 1 out of 925 soil samples.
Voucher material: 13 females, 7 males and 23 juveniles were deposited in the collection of the Museum and Institute of Zoology, Polish Academy of Sciences, Warsaw, Poland under accession numbers as follows: females (MIZ 2/2017/10-15; MIZ 2/2017/29-30; MIZ 2/2017/42-44); males (MIZ 2/2017/16-17; MIZ 2/2017/31-32; MIZ 2/2017/35-36; MIZ 2/2017/45); juveniles (MIZ 2/2017/1-9, MIZ 2/2017/18-26/, MIZ 2/2017/37-41). Five male specimens are kept in collection of the first author (FWK).
Representative DNA sequences: KX137849 (D2-D3 28S rDNA) and KX192397-9, KX192400 (partial 18S-ITS1-partial 5.8S).
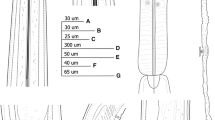
Description (Figs. 1, 2)
Female. [Based on 13 specimens; see also metrical data in Table 1 and Fig. 1A–G). Body C-shaped to open spiral-shaped, more coiled posteriorly. Cuticle with fine, with transverse striations. Cuticle thickness: 3.4–4 at guide ring, 3–4 along most body width, and 7 and 9–10 on ventral and dorsal part of tail. Lips anteriorly flattened to slightly convex, laterally rounded and slightly expanded, set-off from rest of body by almost indistinct to clear constriction. Amphidial fovea pouch-like, protruding to about half of anterior end-guide ring distance, posterior end not bi-lobed. Nerve-ring located at base of odontophore to less than a corresponding body width posterior to it. Pharyngeal bulb occupying about 1/3 of pharynx length. Dorsal and ventro-sublateral gland nuclei 2–2.5 and 3–3.5 wide, respectively. Vagina occupying 46–61 (57.5)% of the corresponding body width, pars distalis and proximalis vaginae 13–15 (4.6) and 15–20 (17.1) long, respectively. Genital tract morphology typical of the genus with sperm cells present in most of examined specimens. Tail bluntly-conoid, ventrally usually flat, sometimes slightly concave or convex; pair of pores present at each side of tail.
Male. [Based on 12 specimens; see also metrical data in Table 1 and Fig. 1H–L). Less frequent than females, sex ratio 1:2. General morphology similar to females, with differences in posterior body part and genital tract. Posterior body part more coiled than in females, tail ventrally concave. Adanal supplements 1–3 pairs, followed by 9–12 single adanal supplements.
Juveniles. [Based on 23 specimens; see also metrical data in Table 2 and Fig. 2A–H). Four developmental stages present. Body from J-shaped to C-shaped in J1, C-shaped in J2, J3 and J4. Shape of lip region in all stages similar to adults, only in J1 lips are not expanded to slightly expanded and not set-off to almost indistinctly set off. Tail shape conoid with a rounded tip in J1 becoming more bluntly rounded in subsequent stages.
Phylogenetic position of L. artemisiae and L. juglandicola
The newly-generated sequences of D2-D3 28S rDNA and sequences containing partial 18S, whole ITS1 region and partial 5.8S are listed together with their GenBank accession numbers in Table 3 and 4. The D2-D3 28S rDNA sequence of the Polish isolate of L. artemisiae was 741 bp long. BLAST search revealed maximum scores with four Russian sequences for L. artemisiae (KF242313–KF242316, 98–99% similarity). Next sequences with highest scores were those for L. uroshis Krnjaić, Lamberti, Krnjaić, Agostinelli & Radicci, 2000 from Slovakia (EF538754, 96% of similarity). The D2-D3 sequence of L. juglandicola was 716 bp long. Results with maximum BLAST scores included a sequence of Longidorus sp. (KF242335) (96% of similarity). These sequences and several other with subsequent highest BLAST scores (listed in Table 3) were used for phylogenetic reconstruction.
In the ITS1 region-containing sequences, four sequence variants were recovered for L. artemisiae and two for L. juglandicola. These sequences were trimmed to include ITS1 only and used for both BLAST search and phylogeny reconstruction. BLAST search of sequences did not show exact matches with sequences of neither L. artemisiae nor L. juglandicola from this study. Sequences of L. artemisiae revealed 80–84% similarity with the sequences for L. elongatus (De Man, 1876) Thorne & Swanger, 1936 and L. intermedius Kozłowska & Seinhorst, 1979 (accession numbers: AF511417, AJ549986–AJ549987, GU199044 and KT308890). Other sequences were characterised by very low query coverage, resulting in low total scores of BLAST searches. None of sequences available in the GenBank database was significantly similar to sequences obtained in this study for the type-population of L. juglandicola from Slovakia. To reconstruct the phylogeny, ten first results from BLAST searches were included to the alignment (some sequences repeated in both searches). The final list of ITS1 sequences used for phylogeny reconstruction is presented in Table 4. The final alignment of the D2-D3 28S rDNA sequences contained 689 positions and the alignment of ITS1 sequences comprised 896 positions. The phylogenetic trees are presented on Fig. 3 A and B. Some relationships were unresolved, however results from both D2-D3 28S rDNA and ITS1 sequences enable two conclusions. First, L. artemisiae as well as L. juglandicola formed their own clades, well distinguishable from the other species in this analysis. Secondly, these two species are not closely related, as many other species appear more closely related to each of them.
Discussion
Morphology and morphometrics of the population of L. artemisiae from Poland studied here revealed several differences in comparison with the original description by Rubtsova et al. (1999). These include a more slender body in the specimens from the type-population (‘a’ index value of 109–155 (133 ± 1.9) and 113–162 (133 ± 1.7) vs 86.6–114.3 (100.8 ± 7.6) and 104.6-133.6 (118.8 ± 9.91) in females and males, respectively. Low ‘a’ index can sometimes be a result of inappropriate preparation of microscopic slides, where specimen is flattened by coverslip. Here microscopic slides were made with care and using glass fibre (see ‘Materials and methods’) Nevertheless, we have prepared additional slides which additional specimens. On these slides pieces of broken coverslip (130–170 µm thick according to the manufacturer) were used instead of glass fibres. Measuring body width of several specimens from such slides did not reveal any important differences compared to values given in Table 1 (data not shown). Other differences worth mentioning include: longer body of male specimens from Poland, 6,201 ± 393.5 vs 5,600 ± 100 µm for the Russian type-population; and longer spicules in specimens of the Polish population compared with the type-population: 39–49 (44 ± 1) vs 47–56 (52 ± 3) µm. Additionally, the population from Poland was characterised by a different sex ratio (1:2 vs 1:1 in the type-population) and the specimens possess lips somewhat less expanded than in the population described by Rubtsova et al. (1999). There was a large difference in odontophore length (see data in Table 1), however this result is not comparable because in the description of Rubtsova et al. (1999) the odontophore was measured from its junction with odontostyle to the posterior end of the ventral sinus while in the present study we measured the entire odontophore.
According to Rubtsova et al. (1999), the code for identifying L. artemisiae in the polytomous key of Chen et al. (1997) is A3-B23-C23-D3-E1-F3-G23-H2-I2. Despite the differences mentioned above, the population of this species from Poland fits into that code and no changes are required. This code is rather similar to that of L. juglandicola which, according to Chen et al. (1997) is A23-B34-C3-D23-E2-F34-G23-H12-I2.
Longidorus artemisiae was described from the rhizosphere of Artemisia sp. (see Rubtsova et al., 1999). Subsequently, Subbotin et al. (2014) reported three populations associated with Elytrigia sp., Poa sp. and Trifolium sp. The present finding of this species in the rhizosphere of nettle in Poland extends the known host plant list of this nematode as well its geographical range.
Similarly to the results obtained by Subbotin et al. (2014), L. artemisiae was most closely related to L. intermedius and L. piceicola based on D2-D3 28S rDNA (Fig. 3A). No comparative data are available for the ITS1 marker of L. artemisiae and any markers of L. juglandicola. The fact that this two species are not closely related was not expected taking into account that L. artemisiae and L. juglandicola are rather similar in terms of general morphology and morphometrics. For example, this similarity is well visible in the code of Chen et al. (1997). Despite that, many other species which are less similar in general morphology, appear to be more closely related to both L. artemisiae and L. juglandicola (Fig. 3A, B).
References
Archidona-Yuste, A., Navas-Cortés, J. A., Cantalapiedra-Navarrete, C., Palomares-Rius, J. E., & Castillo, P. (2016). Unravelling the biodiversity and molecular phylogeny of needle nematodes of the genus Longidorus (Nematoda: Longidoridae) in olive and a description of six new species. PLoS One, 11(1), e0147689.
Barsi, L., & Lamberti, F. (2002). First record of Longidorus juglandicola Lišková et al., 1997 (Nematoda: Dorylaimida) from Serbia. Nematologia Mediterranea, 30, 47–53.
Brown, D. J. F., & Boag, B. (1988). An examination of methods used to extract virus-vector nematodes (Nematoda: Longidoridae and Trichodoridae) from soil samples. Nematologia Mediterranea, 16, 93–99.
Chen, Q. W., Hooper, D. J., Loof, P. A. A., & Xu, J. (1997). A revised polytomous key for the identification of species of the genus Longidorus Micoletzky, 1922 (Nematoda: Dorylaimoidea). Fundamental and Applied Nematology, 20, 15–28.
Cherry, T., Szalanski, A. L., Todd, T. C., & Powers, T. O. (1997). The internal transcribed spacer region of Belonolaimus (Nemata: Belonolaimidae). Journal of Nematology, 29, 23–29.
Courtney, W. D., Polley, D., & Miller, V. L. (1955). TAF, an improved fixative in nematode technique. Plant Disease Reporter, 39, 570–571.
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772.
De Luca, F., Landa, B. B., Mifsud, D., Troccoli, A., Vovlas, N., & Castillo, P. (2009). Molecular characterisation of Longidorus kuiperi Brinkman, Loof & Barbez, 1987 (Nematoda: Longidoridae) from the Mediterranean Basin. Nematology, 11, 155–160.
De Luca, F., Reyes, A., Grunder, J., Kunz, P., Agostinelli, A., De Giorgi, C., & Lamberti, F. (2004). Characterization and sequence variation in the rDNA region of six nematode species of the genus Longidorus (Nematoda). Journal of Nematology, 36, 147–152.
Gutiérrez-Gutiérrez, C., Cantalapiedra-Navarrete, C., Montes-Borrego, M., Palomares-Rius, J. E., & Castillo, P. (2013). Molecular phylogeny of the nematode genus Longidorus (Nematoda: Longidoridae) with description of three new species. Zoological Journal of the Linnean Society, 167, 473–500.
He, Y., Subbotin, S. A., Rubtsova, T. V., Lamberti, F., Brown, D. J. F., & Moens, M. (2005). A molecular phylogenetic approach to Longidoridae (Nematoda: Dorylaimida). Nematology, 7, 111–124.
Kornobis, F. (2012). First record of Longidorus danuvii Barsi, Lamberti and De Luca, 2007 (Nematoda: Longidoridae) from Poland with description of pathologies of the lip region and reproductive system. Helminthologia, 49, 104–107.
Kornobis, F. W., Dobosz, R., Bubniewicz, P., & Filipiak, A. (2016). First record of nematode Longidorus attenuatus on soybean in Poland. Plant Disease,
Kornobis, F. W., & Peneva, V. (2011). Longidorus poessneckensis Altherr, 1974 and L. piceicola Lišková, Robbins & Brown, 1997 (Nematoda: Longidoridae): new records from Poland and the first description of the L. poessneckensis male and a bivulval female. Systematic Parasitology, 80, 205–216.
Krnjaic, D., Lamberti, F., & Radicci, V. (2002). Longidoridae (Nematoda) occurring in the Topchider Park of Belgrade, Serbia, with the description of Paralongidorus serbicus sp. n. Nematologia Mediterranea, 30, 185–200.
Kumari, S., Decraemer, W., Traversa, D., & Lišková, M. (2009). Molecular and morphological delineation of Longidorus poessneckensis Altherr, 1974 (Nematoda: Dorylaimida). European Journal of Plant Pathology, 123, 125–137.
Lišková, M., Robbins, R. T., & Brown, D. J. F. (1997). Descriptions of three new Longidorus species from Slovakia (Nemata: Longidoridae). Journal of Nematology, 29, 336–348.
Maafi, Z. T., Subbotin, S. A., Sturhan, D., Barooti, S., & Taheri, Z. M. (2015). Characterisation of Longidorus iranicus Sturhan & Barooti, 1983 (Nematoda: Longidoridae) from Iran and synonymisation of L. moesicus Lamberti, Choleva & Agostinelli, 1983. Russian Journal of Nematology, 23, 21–28.
Niknam, G., Pedram, M., Nejad, E. G., Ye, W., Robbins, R. T., & Maafi, Z. T. (2010). Morphological and molecular characterisation of Longidorus tabrizicus sp. n. and L. sturhani Rubtsova, Subbotin, Brown and Moens, 2001 (Nematoda: Longidoridae) from north-western Iran. Russian Journal of Nematology, 18, 127–140.
Nunn, G. B. (1992). Nematode molecular evolution. An investigation of evolutionary patterns among nematodes based on DNA sequences. PhD dissertation, Nottingham: University of Nottingham.
Pedram, M., Niknam, G., Robbins, R. T., Decraemer, W., Ye, W., & Yu, Q. (2010). First record of Trichodorus primitivus and morphological and molecular identification of Longidorus elongatus from Canada. Plant Disease, 94, 782.
Peneva, V. K., Lazarova, S. S., De Luca, F., & Brown, D. J. F. (2013). Description of Longidorus cholevae sp. n. (Nematoda, Dorylaimida) from a riparian habitat in the Rila Mountains. Bulgaria. ZooKeys, 330, 1–26.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542.
Roshan-Bakhsh, A., Pourjam, E., & Pedram, M. (2016). Description of Longidorus perangustus sp. n. (Dorylaimida: Longidoridae), an amphimictic species from Iran. European Journal of Plant Pathology, 144, 581–594.
Rubtsova, T. V., Chizhov, V. N., & Subbotin, S. A. (1999). Longidorus artemisiae sp. n. (Nematoda: Longidoridae) from roots of Artemisia sp., Rostov region. Russia. Russian Journal of Nematology, 7, 33–38.
Rubtsova, T. V., Moens, M., & Subbotin, S. A. (2005). PCR amplification of a rRNA gene fragment from formalin-fixed and glycerine-embedded nematodes from permanent slides. Russian Journal of Nematology, 13, 137–140.
Rubtsova, T. V., Subbotin, S. A., Brown, D. J. F., & Moens, M. (2001). Description of Longidorus sturhani sp. n. (Nematoda: Longidoridae) and molecular characterisation of several longidorid species from Western Europe. Russian Journal of Nematology, 9, 127–136.
Seinhorst, J. W. (1959). A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica, 4, 67–69.
Subbotin, S. A., Rogozhin, E. A., & Chizhov, V. N. (2014). Molecular characterisation and diagnostics of some Longidorus species (Nematoda: Longidoridae) from Russia and other countries using rRNA genes. European Journal of Plant Pathology, 138, 377–390.
Subbotin, S. A., Stanley, J. D., Ploeg, A. T., Maafi, Z. T., Tzortzakakis, E. A., Chitambar, J. J., et al. (2015). Characterisation of populations of Longidorus orientalis Loof, 1982 (Nematoda: Dorylaimida) from date palm (Phoenix dactylifera L.) in the USA and other countries and incongruence of phylogenies inferred from ITS1 rRNA and coxI genes. Nematology, 17, 459–477.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Taylor, C., & Brown, D. (1997). Nematode vectors of plant viruses. Wallingford, UK: CAB International.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Tzortzakakis, E. A., Archidona-Yuste, A., Cantalapiedra-Navarrete, C., Nasiou, E., Lazanaki, M. S., Kabourakis, E., et al. (2014). Integrative diagnosis and molecular phylogeny of dagger and needle nematodes of olives and grapevines in the island of Crete, Greece, with description of Xiphinema cretense n. sp. (Nematoda, Longidoridae). European Journal of Plant Pathology, 140, 563–590.
Vrain, T. C., Wakarchuk, D. A., Levesque, A. C., & Hamilton, R. I. (1992). Intraspecific rDNA restriction fragment length (bp) polymorphism in the Xiphinema americanum group. Fundamental and Applied Nematology, 15, 563–573.
Ye, W., Szalanski, A. L., & Robbins, R. T. (2004). Phylogenetic relationships and genetic variation in Longidorus and Xiphinema species (Nematoda: Longidoridae) using ITS1 sequences of nuclear ribosomal DNA. Journal of Nematology, 36, 14–19.
Yoder, M., De Ley, I. T., King, I. W., Mundo-Ocampo, M., Mann, J., Blaxter, M., et al. (2006). DESS: a versatile solution for preserving morphology and extractable DNA of nematodes. Nematology, 8, 367–376.
Funding
Analysis of morphology in this work was carried on the equipment acquired as part of the Innovative Economy Operational Programme conducted by the Museum and Institute of Zoology, Polish Academy of Sciences. Project No. WND-POIG.01.03.01-00-133/09 co-financed by the European Union from the European Regional Development Fund (0.5). The technical support for the realization of this work was also provided within the frame of the project ‘Centrum of Excellence for Parasitology’ No. 26220120022 supported by the operating program ‘Research and Development’ funded by the European Fund for Regional Development (0.4) and project VEGA 2/0013/16 (0.1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed.
Additional information
This article is part of the Topical Collection “Nematoda”.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kornobis, F.W., Renčo, M. & Filipiak, A. First record and description of juvenile stages of Longidorus artemisiae Rubtsova, Chizhov & Subbotin, 1999 (Nematoda: Longidoridae) in Poland and new data on L. juglandicola Lišková, Robbins & Brown, 1997 based on topotype specimens from Slovakia. Syst Parasitol 94, 391–402 (2017). https://doi.org/10.1007/s11230-017-9703-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-017-9703-y