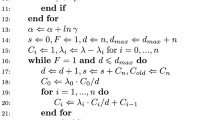Abstract
Hidden Markov models (HMMs) are flexible, well-established models useful in a diverse range of applications. However, one potential limitation of such models lies in their inability to explicitly structure the holding times of each hidden state. Hidden semi-Markov models (HSMMs) are more useful in the latter respect as they incorporate additional temporal structure by explicit modelling of the holding times. However, HSMMs have generally received less attention in the literature, mainly due to their intensive computational requirements. Here a Bayesian implementation of HSMMs is presented. Recursive algorithms are proposed in conjunction with Metropolis-Hastings in such a way as to avoid sampling from the distribution of the hidden state sequence in the MCMC sampler. This provides a computationally tractable estimation framework for HSMMs avoiding the limitations associated with the conventional EM algorithm regarding model flexibility. Performance of the proposed implementation is demonstrated through simulation experiments as well as an illustrative application relating to recurrent failures in a network of underground water pipes where random effects are also included into the HSMM to allow for pipe heterogeneity.




Similar content being viewed by others
References
R Development Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2012)
Baum, L.E., Petrie, T., Soules, G., Weiss, N.: A maximization technique in the statistical analysis of probabilistic functions of Markov chains. Ann. Math. Stat. 41, 164–171 (1970)
Bellone, E., Hughes, J.P., Guttorp, P.: A hidden Markov model for downscaling synoptic atmospheric patterns to precipitation amounts. Clim. Res. 15, 1–12 (2000)
Bulla, J., Bulla, I.: Stylized facts of financial time series and hidden semi-Markov models. Comput. Stat. Data Anal. 51, 2192–2209 (2006)
Bulla, J., Bulla, I., Nenadic, O.: HSMM—an R package for analyzing hidden semi-Markov models. Comput. Stat. Data Anal. 54, 611–619 (2010)
Celeux, G., Hurn, M., Robert, C.P.: Computational and inferential difficulties with mixture posterior distributions. J. Am. Stat. Assoc. 95(451), 957–970 (2000)
Chib, S.: Calculating posterior distributions and modal estimates in Markov mixture models. J. Econom. 75(1), 79–97 (1996)
Devijver, P.A.: Baum’s forward-backward algorithm revisited. Pattern Recognit. Lett. 3(6), 369–373 (1985)
Dewar, M., Wiggins, C., Wood, F.: Inference in hidden Markov models with explicit state duration distributions. IEEE Signal Process. Lett. 19(4), 235–238 (2012)
Dong, M., He, D.: A segmental hidden semi-Markov model (HSMM)-based diagnostics and prognostics framework and methodology. Mech. Syst. Signal Process. 21, 2248–2266 (2007)
Economou, T., Vitolo, R., Bailey, T.C., Kapelan, Z., Waterhouse, E.K.: A latent structure model for high river flows. In: Proceedings of the 24th International Workshop on Statistical Modelling, pp. 125–129 (2009)
Economou, T., Kapelan, Z., Bailey, T.C.: On the prediction of underground water pipe failures: zero-inflation and pipe specific effects. J. Hydroinform. 14(4), 872–883 (2012)
Fearnhead, P., Sherlock, C.: An exact Gibbs sampler for the Markov-modulated Poisson process. J. R. Stat. Soc., Ser. B, Stat. Methodol. 68(5), 767–784 (2006)
Ferguson, J.D.: Variable duration models for speech. In: Ferguson, J.D. (ed.) Proceedings of the Symposium on the Applications of Hidden Markov Models to Text and Speech, Princeton, NJ, pp. 143–179 (1980)
Gamerman, D.: Markov Chain Monte Carlo: Stochastic Simulation for Bayesian Inference. Chapman & Hall, London (1997)
Gelman, A., Roberts, G.O., Gilks, W.R.: Efficient Metropolis jumping rules. Bayesian Stat. 5, 599–607 (1996)
Gelman, A., Carlin, J.B., Stern, H.S., Rubin, D.B.: Bayesian Data Analysis. Chapman & Hall/CRC, London (2004)
Gilks, W.R., Richardson, S., Spiegelhalter, D.J.: Markov Chain Monte Carlo in Practice. Chapman & Hall/CRC, London (1996)
Guedon, Y.: Review of several stochastic speech unit models. Comput. Speech Lang. 6, 377–402 (1992)
Guedon, Y.: Estimating hidden semi-Markov chains from discrete sequences. J. Comput. Graph. Stat. 12(3), 604–639 (2003)
Guha, S., Li, Y., Neuberg, D.: Bayesian hidden Markov modeling of array CGH data. J. Am. Stat. Assoc. 103(482), 485–497 (2008)
Hughes, J.P., Guttorp, P., Charles, S.P.: A non-homogeneous hidden Markov model for precipitation occurrence. J. R. Stat. Soc., Ser. C, Appl. Stat. 48(1), 15–30 (1999)
Jardine, A.K., Lin, D., Banjevic, D.: A review on machinery diagnostics and prognostics implementing condition-based maintenance. Mech. Syst. Signal Process. 20(7), 1483–1510 (2006)
Johnson, M.J., Willsky, A.S.: Bayesian nonparametric hidden semi-Markov models. arXiv:1203.1365v2 (2012)
Jouyaux, C., Richardson, S., Longini, I.: Modeling markers of disease progression by a hidden Markov process: application to characterizing cd4 cell decline. Biometrics 56(3), 733–741 (2000)
Kleiner, Y., Rajani, B.: Comprehensive review of structural deterioration of water mains: statistical models. Urban Water 3, 131–150 (2001)
Kozumi, H.: Bayesian analysis of discrete survival data with a hidden Markov chain. Biometrics 56(4), 1002–1006 (2000)
Levinson, S.E.: Continuously variable duration hidden Markov models for automatic speech recognition. Comput. Speech Lang. 1, 29–45 (1986)
Marin, J.-M., Robert, C.P.: Bayesian Core: A Practical Approach to Computational Bayesian Statistics. Springer, Berlin (1997)
Rabiner, L.: A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 77(2), 257–285 (1989)
Richardson, S., Green, P.J.: On Bayesian analysis of mixtures with an unknown number of components. J. R. Stat. Soc. B 59(4), 731–792 (1997)
Robert, C.P., Titterington, D.M.: Reparameterization strategies for hidden Markov models and Bayesian approaches to maximum likelihood estimation. Stat. Comput. 8, 145–158 (1998)
Robert, C.P., Rydén, T., Titterington, D.M.: Bayesian inference in hidden Markov models through the reversible jump Markov chain Monte Carlo method. J. R. Stat. Soc. B 62(1), 57–65 (2000)
Rydén, T., Terasvirta, T., Asbrink, S.: Stylized facts of daily return series and the hidden Markov model. J. Appl. Econom. 13, 217–244 (1998)
Sansom, J., Thomson, P.: Fitting hidden semi-Markov models to breakpoint rainfall data. J. Appl. Probab. 38A, 142–157 (2001)
Schmidler, S.C., Liu, J.S., Brutlag, D.L.: Bayesian segmentation of protein secondary structure. J. Comput. Biol. 7(1–2), 233–248 (2000)
Scott, S.: Bayesian methods for hidden Markov models: recursive computing in the 21st century. J. Am. Stat. Assoc. 97, 337–351 (2002)
Scott, S., Smyth, P.: The Markov modulated Poisson process and Markov Poisson cascade with applications to web traffic modelling. Bayesian Stat. 7, 671–680 (2003)
Spiegelhalter, D.J., Best, N.G., Carlin, B.P., Van Der Linde, A.: Bayesian measures of model complexity and fit. J. R. Stat. Soc., Ser. B, Stat. Methodol. 64(4), 583–639 (2002)
Stephens, M.: Dealing with label switching in mixture models. J. R. Stat. Soc. B 62(4), 795–809 (2000)
Tokdar, S., Xi, P., Kelly, R., Kass, R.: Detection of bursts in extracellular spike trains using hidden semi-Markov point process models. J. Comput. Neurosci. 29, 203–212 (2010)
Yau, C., Papaspiliopoulos, O., Roberts, G.O., Holmes, C.: Bayesian non-parametric hidden Markov models with applications in genomics. J. R. Stat. Soc. B 73(1), 37–57 (2011)
Yu, S.-Z.: Hidden semi-Markov models. Artif. Intell. 174, 215–243 (2010)
Acknowledgements
The pipe dataset used in this paper was provided by Dr. Yehuda Kleiner whom the authors gratefully acknowledge.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A: Pseudo code for simulation experiment with Gaussian observations
Simulate data from 2-state Gaussian HSMM, Eq. (10):
-
1.
Set values for μ S ,σ,ϕ S and π S for S=1,2.
-
2.
Set i=1 and sample initial state S i from π={π S }.
-
3.
Sample duration τ i of S i , from zero-truncated Poisson with parameter \(\phi_{S_{i}}\).
-
4.
Sample state transition from distribution corresponding to \(S_{1}^{\mathrm{th}}\) row of P.
-
5.
Repeat steps 3 and 4 until ∑ i τ i ≥250.
-
6.
If ∑ i τ i >250, truncate so that ∑ i τ i =250.
-
7.
For each T=1,…,250, sample from N(μ S ,σ 2) according to which state S the chain is at time step T.
Metropolis-Hastings:
-
Set i=0 and initialise parameters \(\mu_{S}^{(i)}, \sigma^{(i)}, \phi _{S}^{(i)}\) and \(\pi_{S}^{(i)}\).
-
Calculate ℓ (i), the log-likelihood using the forward algorithm in Sect. 3.2.
-
Calculate P (i), the log-posterior by
 (15)
(15)where p(⋅) is the prior for each parameter (Table 1).
Do for i=1,…,M where M is the required number of MCMC iterations:
- μ S ::
-
\(\mu_{S}^{\ast}= \mu_{S}^{(i-1)} + \varepsilon\), \(\varepsilon\sim\mbox {N}(0,\sigma^{2}_{\mu})\)
Calculate ℓ ∗ and hence P ∗ using Eq. (15)
Calculate acceptance probability as η=min(1,Ω) where Ω=exp{P ∗−P (i−1)}
Sample U from U(0,1) If U≤η, set \(\mu_{S}^{(i)}=\mu_{S}^{\ast}\), ℓ (i)=ℓ ∗ and P (i)=P ∗.
- σ::
-
σ ∗=exp{log(σ (i−1))+ε}, \(\varepsilon \sim\mbox{N}(0,\sigma^{2}_{\sigma})\)
Calculate ℓ ∗ and hence P ∗ using Eq. (15)
Calculate acceptance probability as η=min(1,Ω) where Ω=exp{P ∗+log(σ ∗)−[P (i−1)+log(σ (i−1))]}
Sample U from U(0,1)
If U≤η, set σ (i)=σ ∗, ℓ (i)=ℓ ∗ and P (i)=P ∗.
- ϕ S :
-
Same as σ, replacing ϕ S with σ.
- π S :
-
Sample π ∗∼Dir(απ (i−1))
Calculate ℓ ∗ and hence P ∗ using Eq. (15)
Calculate acceptance probability as η=min(1,Ω) where

where d(π;θ) is a Dirichlet density with parameter θ
Sample U from U(0,1)
If U≤η, set \(\pi_{S}^{(i)}=\pi_{S}^{\ast}\), ℓ (i)=ℓ ∗ and P (i)=P ∗.
Adjust \(\sigma^{2}_{\mu}, \sigma^{2}_{\sigma}, \sigma^{2}_{\phi}\) and α to achieve desired acceptance rates.
Note that R code for implementing the HSMMs and HMMs from both simulation experiments is available both as supplementary material to the paper and on http://empslocal.ex.ac.uk/people/staff/te201/HSMM/.
Appendix B: Pseudo code for simulation experiment with NHPP observations
Simulate data from 3-state NHPP-HSMM with intensity function λ(t;x|S) as in Eq. (12), for 50 hypothetical objects:
-
1.
Set values for θ S ,β 0,β 1,ϕ S ,π S and P for S=1,2,3.
-
2.
Sample B j ∼U(80,150), for j=1,…,50, to decide the observation period in discrete time steps for each object.
-
3.
Sample x j ∼U(50,150) to set values for the covariate x.
-
4.
Set j=1, i=1 and sample initial state S i from π={π S }.
-
5.
Sample duration τ i of S i , from zero-truncated Poisson with parameter \(\phi_{S_{i}}\).
-
6.
Sample state transition from distribution corresponding to \(S_{1}^{\mathrm{th}}\) row of P.
-
7.
Repeat steps 3 and 4 until ∑ i τ i ≥B j .
-
8.
If ∑ i τ i >B j , truncate so that ∑ i τ i =B j .
-
9.
For each T=1,…,B j , sample from Pois(Λ([T−1,T]|S)) according to which state S the chain is at time step T, where \(\varLambda([T-1,T]|S)=\int_{T-1}^{T} \lambda(t;x|S) dt\).
-
10.
Repeat steps 4–9 for j=2,…,50.
Metropolis-Hastings:
-
Very similar to the one in Appendix A.
-
The forward algorithm needs to be ran for each of the j=1,…,50 objects so that the log-likelihood \(\ell^{(i)} = \sum_{j=1}^{50} \ell_{j}^{(i)}\) (assuming independence of the objects).
-
The log-posterior is obtained essentially as in Eq. (15). However, we restrict the posterior to be zero in areas of the parameter space other than θ 1<θ 2<θ 3 to impose the label-switching measure.
-
Strictly positive parameters such as θ S and ϕ S are updated the same as σ and ϕ S in Appendix A.
-
Parameter π and vectors made up of rows from transition matrix P, excluding zero elements, are updated exactly like π in Appendix A.
Rights and permissions
About this article
Cite this article
Economou, T., Bailey, T.C. & Kapelan, Z. MCMC implementation for Bayesian hidden semi-Markov models with illustrative applications. Stat Comput 24, 739–752 (2014). https://doi.org/10.1007/s11222-013-9399-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11222-013-9399-z