Abstract
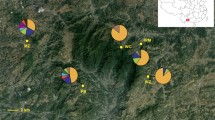
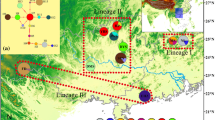
Trailliaedoxa gracilis W. W. Smith et Forrest (Rubiaceae), a Chinese endemic monotypic genus belonging to the Alberteae (Rubiaceae), exhibits a narrow distribution in the dry valleys of the Jinsha River and Red River drainage area in southwestern China. The few sites at which T. gracilis occurs are fragmented and isolated, and several are highly vulnerable to human disturbance. As T. gracilis is a protected plant with a second-degree national priority, the genetic diversity and structure of the populations of this species should be investigated to determine the most suitable conservation strategy. In this study, two chloroplast regions and one nuclear region were used to investigate the genetic diversity, genetic structure, and demographic history of T. gracilis. We observed a high total genetic diversity (H T = 0.952 and 0.966) and low average within-population diversity (H S = 0.07 and 0.489) based on cpDNA and nDNA analyses. Thus, a strong genetic structure (F ST = 0.98049 and 0.59731) was detected. A phylogeographic structure was detected by nuclear DNA analysis (N ST > G ST, P < 0.05); however, the chloroplast data did not show a significant phylogeographic structure (N ST < G ST, P > 0.05). The Bayesian skyline plot and isolation with migration analysis were used to estimate the demographic history of T. gracilis. The results indicated that a marked bottleneck effect occurred during the glacial-interglacial of the Pleistocene. Among the extant populations of T. gracilis, the population found in ChunJiang, LuQuan, and YuXi showed the highest haplotype diversity based on cpDNA sequences and should be given priority for protection. According to the nDNA analysis, every population presented a high level of diversity and a high content of private haplotypes. Therefore, every population should be protected.






Similar content being viewed by others
References
Avise JC, Hamrick JL (1996) Conservation genetics, case histories from nature. Chapman & Hall, New York
Chiang TY, Chiang YC, Chen Y, Chou CH, Havanond S, Hong T, Huang S (2001) Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Mol Ecol 10:2697–2710
Clark AG (1990) Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol 7:111–122
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134:959–969
Doyle J (1991) DNA protocols for plants—CTAB total DNA isolation. Molecular techniques in taxonomy. Springer, Berlin
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:1–8. doi:10.1186/1471-2148-7-214
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Echt CS, DeVerno LL, Anzidei M, Vendramin GG (1998) Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa. Ait Mol Ecol 7:307–316
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinforma 1:47–50
Frankel OH, Brown AHD, Burdon JJ (1995) The conservation of plant biodiversity. Cambridge University Press, Cambridge
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Gong X, Luan SS, Hung KH, Hwang CC, Lin CJ, Chiang YC, Chiang TY (2011) Population structure of Nouelia insignis (Asteraceae), an endangered species in southwestern China, based on chloroplast DNA sequences: recent demographic shrinking. J Plant Res 124:221–230
Graur D, Li WH (2000) Fundamentals of molecular evolution, 2nd edn. Sinauer Associates, Sunderland
Guan ZT, Zhou L (1996) Cycads of China. Sichuan Science and Technology Press, Chengdu
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series. 95–98
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467
Harpending H (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Harrison S, Yu G, Takahara H, Prentice I (2001) Palaeovegetation (communications arising): diversity of temperate plants in East Asia. Nature 413:129–130
Hey J, Nielsen R (2004) Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167:747–760
Hey J, Nielsen R (2007) Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci 104:2785
Hijmans R, Cruz M, Rojas E, Guarino L (2001) DIVA-GIS version 1.4: a geographic information system for the analysis of biodiversity data, manual
Huang S, Chiang YC, Schaal BA, Chou CH, Chiang TY (2001) Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Mol Ecol 10:2669–2681
Jakob SS, Blattner FR (2006) A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol Biol Evol 23:1602–1612
Jia J, Wu H, Wang JF, Gong X (2014) Genetic diversity and structure of Munronia delavayi Franch. (Meliaceae), an endemic species in the dry-hot valley of Jinsha River, south-western China. Genet Resour Crop Evol 61:1381–1395
Jin ZZ (1998) Study on the floristic elements of seed plant in the dry-warm valleys of Yunnan and Sichuan. Guihaia 18:313–321
Jin ZZ (1999) The floristic study on seed plants in the dry-hot valleys in Yunnan and Sichuan. Guihaia 19:1–14
Jin ZZ (2002) Floristic features of dry-hot and dry-warm valleys, Yunnan and Sichuan. Yunnan Science & Technology Press, Yunnan
Kainulainen K, Razafimandimbison SG, Bremer B (2013) Phylogenetic relationships and new tribal delimitations in subfamily Ixoroideae (Rubiaceae). Bot J Linn Soc 173:387–406
Kelchner SA (2000) The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann Mo Bot Gard 87:482–498
Kruckeberg AR, Rabinowitz D (1985) Biological aspects of endemism in higher plants. Annu Rev Ecol Syst 16:447–479
Léotard G, Duputié A, Kjellberg F, Douzery EJ, Debain C, de Granville JJ, McKey D (2009) Phylogeography and the origin of cassava: new insights from the northern rim of the Amazonian basin. Mol Phylogenet Evol 53:329–334
Li RN, Du F, Ma M, Liu Y, Liu J, Liu CY (2012) Study on distribution characteristics of national key protected wild plants in Northwest Yunnan. J West China For Sci 42:53–59
Luan S, Chiang T-Y, Gong X (2006) High genetic diversity vs. low genetic differentiation in Nouelia insignis (Asteraceae), a narrowly distributed and endemic species in China, revealed by ISSR fingerprinting. Ann Bot 98:583–589
Luo XR, Gao YZ, Chen WQ, Ruan YZ (1999) Flora Reipublicae Popularis Sinicae, vol 71. Science Press, Beijing
Luo Z, Diao Y, Yang L, Peng J, Liu Q, Chen F (2009) Community structure of Jatropha curcas in the Jinsha River dry-hot valley in Liangshan Prefecture, Sichuan, China. Chin J Appl Environ Microbiol 15:432–436
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Olsen KM (2002) Population history of Manihot esculenta (Euphorbiaceae) inferred from nuclear DNA sequences. Mol Ecol 11:901–911
Olsen KM, Schaal BA (1999) Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proc Natl Acad Sci 96:5586–5591
Ou XK (1994) The plant resources and its ecological characteristics in dry-hot valley of Jinsha River. J Plant Resour Environ 3:42–46
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Petit RJ, Pineau E, Demesure B, Bacilieri R, Ducousso A, Kremer A (1997) Chloroplast DNA footprints of postglacial recolonization by oaks. Proc Natl Acad Sci 94:9996–10001
Pogson GH, Taggart CT, Mesa KA, Boutilier RG (2001) Isolation by distance in the Atlantic cod, Gadus morhua, at large and small geographic scales. Evolution 55:131–146
Pons O, Petit R (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Rambaut A, Drummond A (2007) Tracer: MCMC trace analysis tool, version 1.5. Tracer website
Ribeiro R, Lemos-Filho J, Ramos A, Lovato M (2010) Phylogeography of the endangered rosewood Dalbergia nigra (Fabaceae): insights into the evolutionary history and conservation of the Brazilian Atlantic Forest. Heredity 106:46–57
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Royden LH, Burchfiel BC, van der Hilst RD (2008) The geological evolution of the Tibetan Plateau. Science 321:1054–1058
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Shaw J et al (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288
Strand A, Leebens Mack J, Milligan B (1997) Nuclear DNA‐based markers for plant evolutionary biology. Mol Ecol 6:113–118
Svejda B et al (2010) Anticancer activity of novel plant extracts from Trailliaedoxa gracilis (WW Smith & Forrest) in human carcinoid KRJ-I cells. Anticancer Res 30:55–64
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Templeton AR, Sing CF (1993) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics 134:659–669
Wang ZH, Liu ED, Xiang CL, Peng H (2011) Revised description and new distribution report of Trailliaedoxa gracilis (Rubiaceae), an endemic species of China. Guihaia 31:569–571
Woerner AE, Cox MP, Hammer MF (2007) Recombination-filtered genomic datasets by information maximization. Bioinformatics 23:1851–1853
Wolfe KH, Li W-H, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci 84:9054–9058
Wright S (1978) Evolution and the genetics of populations. Variability within and among natural populations. University of Chicago Press, Chicago
Wu H, Zeng LQ, Gong X (2010) Chromosome number of Trailliaedoxa gracilis (Rubiaceae) endemic to Jinsha River valley. Acta Bot Yunnanica 5:407–408
Yan FL (1984) Natural geography of China. Science Press, Beijing
Zhan QQ, Wang JF, Gong X, Peng H (2011) Patterns of chloroplast DNA variation in Cycas debaoensis (Cycadaceae): conservation implications. Conserv Genet 12:959–970
Zhang Q, Liu Y, Sodmergen (2003) Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol 44:941–951
Zhao YJ, Gong X (2012) Genetic structure of the endangered Leucomeris decora (Asteraceae) in China inferred from chloroplast and nuclear DNA markers. Conserv Genet 13:271–281
Zhong XH (2000) Degradation of ecosystem and ways of its rehabilitation and reconstruction in dry and hot valley—take representative area of Jinsha River, Yunnan Province as an example. Resources and Environment in the Yangtze Basin 3:376–383
Zhou SZ, Wang XL, Wang J, Xu LB (2006) A preliminary study on timing of the oldest Pleistocene glaciation in Qinghai–Tibetan. Plateau Quatern Int 154:44–51
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (30870242).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jia, J., Zeng, L. & Gong, X. High Genetic Diversity and Population Differentiation in the Critically Endangered Plant Species Trailliaedoxa gracilis (Rubiaceae). Plant Mol Biol Rep 34, 327–338 (2016). https://doi.org/10.1007/s11105-015-0924-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0924-4