Abstract
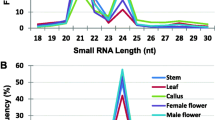
Deep sequencing has advanced the discovery and analysis of the small RNA component of transcriptomes and has revealed developmentally-regulated populations of small RNAs consistent with key roles in plant development. To study small RNA transcriptome complexity and explore their roles in sugarcane development, we obtained almost 50 million small RNA reads from suspension cells, embryogenic calli, leaf, apex and a developmental series of stem internodes. The complexity of the small RNA component of the transcriptome varied between tissues. The undifferentiated and young tissue type libraries had lower redundancy levels than libraries generated from maturing and mature tissues. The ratio of 21:24 nt small RNAs also varied widely between different tissue types, as did the proportion of abundant small RNAs derived from each putative origin of small RNA biogenesis. Cluster analysis indicates many abundant small RNAs display developmental expression patterns. There was substantial variation in isomiR composition, abundance and expression patterns within sugarcane microRNA (miRNA) families. Two hundred and fifty-six isomiRs from 36 miRNA families were identified by homology to known miRNA families from a range of plant species. Many isomiRs and miRNA families appear to be developmentally regulated, including a subset of miRNAs that are progressively upregulated during stem internode maturation. Transcribed sequences putatively targeted by abundant sugarcane small RNAs were predicted and miRNA directed cleavage of 18 predicted sugarcane targets were validated by 5′ RACE.









Similar content being viewed by others
References
Arruda P (2012) Genetically modified sugarcane for bioenergy generation. Curr Opin Biotechnol 23:315–322. doi:10.1016/j.copbio.2011.10.012
Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K (2004) Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res 11:93–99
Basnayake SW, Moyle R, Birch RG (2011) Embryogenic callus proliferation and regeneration conditions for genetic transformation of diverse sugarcane cultivars. Plant Cell Rep 30:439–448. doi:10.1007/s00299-010-0927-4
Bottino MC et al (2013) High-throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS One 8:e59423. doi:10.1371/journal.pone.0059423
Calsa Junior T, Carraro DM, Benatti MR, Barbosa AC, Kitajima JP, Carrer H (2004) Structural features and transcript-editing analysis of sugarcane (Saccharum officinarum L.) chloroplast genome. Curr Genet 46:366–373
Casu RE, Grof CP, Rae AL, McIntyre CL, Dimmock CM, Manners JM (2003) Identification of a novel sugar transporter homologue strongly expressed in maturing stem vascular tissues of sugarcane by expressed sequence tag and microarray analysis. Plant Mol Biol 52:371–386
Chen CJ, Liu Q, Zhang YC, Qu LH, Chen YQ, Gautheret D (2011) Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biol 8:538–547. doi:10.4161/rna.8.3.15199
Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH (2010a) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107:15269–15274
Chen X (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579:5923–5931. doi:10.1016/j.febslet.2005.07.071
Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25:21–44. doi:10.1146/annurev.cellbio.042308.113417
Chen X (2012) Small RNAs in development—insights from plants. Curr Opin Genet Dev 22:361–367. doi:10.1016/j.gde.2012.04.004
Chen X, Zhang Z, Liu D, Zhang K, Li A, Mao L (2010b) SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J Integr Plant Biol 52:946–951. doi:10.1111/j.1744-7909.2010.00987.x
Chou TC, Moyle RL (2014) Synthetic versions of firefly luciferase and Renilla luciferase reporter genes that resist transgene silencing in sugarcane. BMC Plant Biol 14:92. doi:10.1186/1471-2229-14-92
D'Hont A, Grivet L, Feldmann P, Rao S, Berding N, Glaszmann JC (1996) Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol Gen Genet 250:405–413
Damaj MB et al (2010) Sugarcane DIRIGENT and O-methyltransferase promoters confer stem-regulated gene expression in diverse monocots. Planta 231:1439–1458. doi:10.1007/s00425-010-1138-5
de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20:1453–1454. doi:10.1093/bioinformatics/bth078
Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF (2012) Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 8:e1002419
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Ferreira TH, Gentile A, Vilela RD, Costa GG, Dias LI, Endres L, Menossi M (2012) microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS One 7:e46703
Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N (2008) Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26:407–415. doi:10.1038/Nbt1394
Gentile A et al (2013) Effects of drought on the microtranscriptome of field-grown sugarcane plants. Planta 237:783–798. doi:10.1007/s00425-012-1795-7
Hoen PA et al (2008) Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36:e141
Jackson MA, Anderson DJ, Birch RG (2013) Comparison of Agrobacterium and particle bombardment using whole plasmid or minimal cassette for production of high-expressing, low-copy transgenic plants. Transgenic Res 22:143–151
Jackson MA, Sternes PR, Mudge SR, Graham MW, Birch RG (2014) Design rules for efficient expression in plants. Plant Biotechnol J 12:925–933. doi:10.1111/pbi.12197
Kinkema M et al (2014) Improved molecular tools for sugar cane biotechnology. Plant Mol Biol 84:497–508. doi:10.1007/s11103-013-0147-8
Koia J, Moyle R, Hendry C, Lim L, Botella JR (2013) Pineapple translation factor SUI1 and ribosomal protein L36 promoters drive constitutive transgene expression patterns in Arabidopsis thaliana. Plant Mol Biol 81:327–336
Koia JH, Moyle RL, Botella JR (2012) Microarray analysis of gene expression profiles in ripening pineapple fruits BMC. Plant Biol 12:240. doi:10.1186/1471-2229-12-240
Kuang H et al (2009) Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Res 19:42–56. doi:10.1101/gr.078196.108
Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52:133–146
Mackowiak SD (2011) Identification of novel and known miRNAs in deep-sequencing data with miRDeep2. Curr Protoc Bioinformatics. doi:10.1002/0471250953.bi1210s36, Chapter 12: Unit 12 10
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Mendes R, Pizzirani-Kleiner AA, Araujo WL, Raaijmakers JM (2007) Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl Environ Microbiol 73:7259–7267. doi:10.1128/AEM.01222-07
Morin RD et al (2008) Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18:610–621. doi:10.1101/gr.7179508
Moyle R, Fairbairn DJ, Ripi J, Crowe M, Botella JR (2005) Developing pineapple fruit has a small transcriptome dominated by metallothionein. J Exp Bot 56:101–112. doi:10.1093/jxb/eri015
Moyle RL, Birch RG (2013a) Diversity of sequences and expression patterns among alleles of a sugarcane loading stem gene. Theor Appl Genet 126:1775–1782. doi:10.1007/s00122-013-2091-z
Moyle RL, Birch RG (2013b) Sugarcane loading stem gene promoters drive transgene expression preferentially in the stem. Plant Mol Biol 82:51–58. doi:10.1007/s11103-013-0034-3
Moyle RL, Sternes PR, Birch RG (2014) Incorporating Target Sequences of Developmentally-Regulated Small RNAs into Transgenes to Enhance Tissue Specificity of Expression in Plants Unpublished manuscript under review at Plant Mol Biol Rep
Mudge SR, Basnayake SW, Moyle RL, Osabe K, Graham MW, Morgan TE, Birch RG (2013) Mature-stem expression of a silencing-resistant sucrose isomerase gene drives isomaltulose accumulation to high levels in sugarcane. Plant Biotechnol J 11:502–509. doi:10.1111/pbi.12038
Ortiz-Morea FA, Vicentini R, Silva GF, Silva EM, Carrer H, Rodrigues AP, Nogueira FT (2013) Global analysis of the sugarcane microtranscriptome reveals a unique composition of small RNAs associated with axillary bud outgrowth. J Exp Bot 64:2307–2320. doi:10.1093/jxb/ert089
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263. doi:10.1038/nature01958
Piriyapongsa J, Jordan IK (2008) Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 14:814–821. doi:10.1261/rna.916708
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Ruwe H, Schmitz-Linneweber C (2012) Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res 40:3106–3116. doi:10.1093/nar/gkr1138
Shen D, Wang S, Chen H, Zhu QH, Helliwell C, Fan LJ (2009) Molecular phylogeny of miR390-guided trans-acting siRNA genes (TAS3) in the grass family. Plant Syst Evol 283:125–132. doi:10.1007/s00606-009-0221-5
Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM (2007) Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134:1051–1060
Solomon EI, Sundaram UM, Machonkin TE (1996) Multicopper oxidases and oxygenases. Chem Rev 96:2563–2606
Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128:1127–1135
Taylor PWJ, Ko HL, Adkins SW, Rathus C, Birch RG (1992) Establishment of embryogenic callus and high protoplast yielding suspension-cultures of sugarcane (Saccharum spp. hybrids). Plant Cell Tissue Organ Cult 28:69–78. doi:10.1007/bf00039917
Thiebaut F et al (2012) Computational identification and analysis of novel sugarcane microRNAs. BMC Genomics 13:290. doi:10.1186/1471-2164-13-290
Van Dillewijn C (1952) Botany of sugarcane. Chronica Botanica Co, Waltham
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wang X et al (2009) Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21:1053–1069. doi:10.1105/tpc.109.065714
Wu B, Wang M, Ma Y, Yuan L, Lu S (2012) High-throughput sequencing and characterization of the small RNA transcriptome reveal features of novel and conserved microRNAs in Panax ginseng. PLoS One 7:e44385. doi:10.1371/journal.pone.0044385
Yan Y, Zhang Y, Yang K, Sun Z, Fu Y, Chen X, Fang R (2011) Small RNAs from MITE-derived stem-loop precursors regulate abscisic acid signaling and abiotic stress responses in rice. Plant J 65:820–828. doi:10.1111/j.1365-313X.2010.04467.x
Zabala G et al (2012) Divergent patterns of endogenous small RNA populations from seed and vegetative tissues of Glycine max. BMC Plant Biol 12:177. doi:10.1186/1471-2229-12-177
Zanca AS, Vicentini R, Ortiz-Morea FA, Del Bem LE, da Silva MJ, Vincentz M, Nogueira FT (2010) Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane. BMC Plant Biol 10:260. doi:10.1186/1471-2229-10-260
Zhang L, Zheng Y, Jagadeeswaran G, Li Y, Gowdu K, Sunkar R (2011) Identification and temporal expression analysis of conserved and novel microRNAs in Sorghum. Genomics 98:460–468. doi:10.1016/j.ygeno.2011.08.005
Zhao YT, Wang M, Fu SX, Yang WC, Qi CK, Wang XJ (2012) Small RNA profiling in two Brassica napus cultivars identifies microRNAs with oil production- and development-correlated expression and new small RNA classes. Plant Physiol 158:813–823. doi:10.1104/pp.111.187666
Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9:149
Acknowledgments
We thank Daniel Barry for technical assistance, Michael Graham for assistance with calli and suspension cell library data acquisition and Robert Birch for assistance with project conception and editing of the manuscript. This research was supported through a collaboration between CSR Sugar Limited (Sucrogen) and The University of Queensland under the Australian Research Council’s Linkage scheme.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file 1
(XLSX 27 kb)
Supplementary file 5
(XLSX 353 kb)
Supplementary file 6
(XLSX 11 kb)
Rights and permissions
About this article
Cite this article
Sternes, P.R., Moyle, R.L. Deep Sequencing Reveals Divergent Expression Patterns Within the Small RNA Transcriptomes of Cultured and Vegetative Tissues of Sugarcane. Plant Mol Biol Rep 33, 931–951 (2015). https://doi.org/10.1007/s11105-014-0787-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0787-0