Abstract
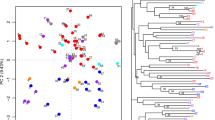
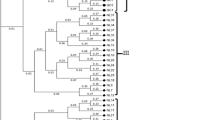
Cranberries (Vaccinium macrocarpon Ait.) are an economically important fruit crop derived from a North American native species. We report the application of 12 simple sequence repeats (SSR) or microsatellite markers to assess the genetic diversity of cranberry cultivars. We studied 164 samples of 21 different cranberry cultivars, 11 experimental hybrids, and 6 representative accessions of wild species. Genetic cluster analysis, based on 117 SSR alleles, differentiated the major cranberry cultivars. However, some cranberry cultivar subclone variants and mislabeled samples were observed. Consensus genetic profiles identified the most likely clonal representatives of several important cranberry cultivars (e.g., “Ben Lear,” “Howes,” and “Stevens”). The markers were further used to confirm putative parents of several hybrid progenies. The long-term goal of our studies is to identify, preserve, and utilize unique genetic materials to breed improved cranberries. Attaining this goal will help growers maintain sustainability under changing economic and environmental conditions.


Similar content being viewed by others
References
Areškevičiūtė J, Paulauskas A, Česonienė L, Daubaras R (2006) Genetic characterisation of wild cranberry (Vaccinium oxycoccos) from Čepkeliai reserve by the RAPD method. Biologija 1:5–7
Caruso FL, Bristow PR, Oudemans PV (2000) Cranberries: the most intriguing native North American fruit. APSnet Features. doi:10.1094/APSnetFeature-2000-1100
Erfani J, Ebadi A, Abdollahi H, Fatahi R (2012) Genetic diversity of some pear cultivars and genotypes using simple sequence repeat (SSR) markers. Plant Mol Biol Rep. doi:10.1007/s11105-012-0421-y
Georgi L, Herai RH, Vidal R, Carazzolle MF, Pereira GG, Polashock J, Vorsa N (2012) Cranberry microsatellite marker development from assembled next-generation genomic sequence. Mol Breed. doi:10.1007/s11032-011-9613-7
Novy RG, Vorsa N (1994) DNA fingerprinting of cranberry varieties using RAPDs. In: Roper TR (ed) Wisconsin Cranberry School 1994 proceedings.
Novy RG, Vorsa N (1995) Identification of intracultivar genetic heterogeneity in cranberry using silver-stained RAPDs. HortScience 30:600–604
Novy RG, Vorsa N (1996) Evidence for RAPD heteroduplex formation in cranberry: Implications for pedigree and genetic relatedness studies and a source of codominant RAPD markers. Theor Appl Genet 92:840–849
Novy RG, Kobak C, Goffreda J, Vorsa N (1994) RAPDs identify varietal misclassification and regional divergence in cranberry (Vaccinium macrocarpon Ait.). Theor Appl Genet 88:1004–1010
Novy RG, Vorsa N, Patten K (1996) Identifying genotypic heterogeneity in McFarlin’ cranberry: a randomly-amplified polymorphic DNA (RAPD) and phenotypic analysis. HortScience 2:210–215
Palombi M, Damiano C (2002) Comparison between RAPD and SSR molecular markers in detecting genetic variation in kiwifruit (Actinidia deliciosa A. Chev). Plant Cell Rep 20:1061–1066
Panwar P, Nath M, Yadav VK, Kumar A (2010) Comparative evaluation of genetic diversity using RAPD, SSR and cytochrome P450 gene based markers with respect to calcium content in finger millet (Eleusine coracana L. Gaertn.). J Genet 89:121–133
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Polashock J, Vorsa N (2002) Development of SCAR markers for DNA fingerprinting and germplasm analysis of American cranberry. HortScience 127:677–687
Potts SM, Yuepeng H, Khan MA, Kushad MM, Rayburn AL, Korban SS (2012) Genetic diversity and characterization of a core collection of Malus germplasm using simple sequence repeats (SSRs). Plant Mol Biol Rep 30:827–837
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ravi M, Geethanjali S, Sameeyafarheen F, Maheswaran M (2003) Molecular marker based genetic diversity analysis in rice (Oryza sativa L.) using RAPD and SSR markers. Euphytica 133:243–252
Roper TR (2008) Cranberry production in Wisconsin. http://www.wiscran.org/user_image/pdf_files/CranProduction08.pdf.
Roper TR, Skroch P, Nienhuis J (1995) Barren berry and Searles vines are genetically dissimilar. Cranberries 59:14–15
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. J Trends Biotech 23:48–55
Zhang Q, Li J, Zhao Y, Korban SS, Han Y (2012) Evaluation of genetic diversity in Chinese wild apple species along with apple cultivars using SSR markers. Plant Mol Biol Rep 30:539–546
Zhu H, Senalik D, McCown BH, Zeldin EL, Speers J, Hyman J, Bassil N, Hummer K, Simon PW, Zalapa JE (2012) Mining and validation of pyrosequenced simple sequence repeats (SSRs) from American cranberry (Vaccinium macrocarpon Ait.). Theor Appl Genet 124:87–96
Acknowledgments
The authors thank PS100, Emily Gustin, and Eric Wiesman for their help with different aspects of this manuscript. B. McCown, E. Zeldin, E. Grygleski, and Rod Serres provided cranberry samples. This project was supported by the Integrative Biological Sciences Summer Research Program at the University of Wisconsin–Madison through a National Foundation Science grant (DBI-1063085), the Wisconsin Cranberry Growers Association, and the USDA-ARS (project number 3655-21220-001-00).
Author information
Authors and Affiliations
Corresponding author
Additional information
D. Fajardo, J. Morales, and H. Zhu contributed equally to the work described in this manuscript.
Rights and permissions
About this article
Cite this article
Fajardo, D., Morales, J., Zhu, H. et al. Discrimination of American Cranberry Cultivars and Assessment of Clonal Heterogeneity Using Microsatellite Markers. Plant Mol Biol Rep 31, 264–271 (2013). https://doi.org/10.1007/s11105-012-0497-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-012-0497-4